Christine Gronseth, CG(ASCP)CM;
Scott McElhone, MB(ASCP)CM; Bart Scott, MD;
Cecilia Yeung, MD; Min Fang, MD, PhD
 CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from Seattle Cancer Care Alliance. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from Seattle Cancer Care Alliance. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
November 2016—Copy neutral loss of heterozygosity (cnLOH) is an acquired abnormality found in patients with cancer and hematologic disorders and can be detected by molecular techniques such as PCR-based analyses and hybridization-based chromosome genomic array testing (CGAT). We report a case in which cnLOH was the sole abnormality detected by CGAT in a patient with myelodysplastic syndrome. This case illuminates the importance of utilizing CGAT results, namely cnLOH findings, as one of the primary diagnostic indicators in order to expedite initial therapies.
Introduction. The 2008 World Health Organization criteria recognize the initial diagnosis of MDS as cytopenia of undetermined origin in the blood and greater than five percent blasts in the bone marrow, or less than 10 percent blasts in the bone marrow if unequivocal dysplasia is present along with a cytogenetic abnormality of: -5 or del(5q), -7 or del(7q), del(9q), del(11q), del(12p) or t(12p), -13 or del(13q), i(17q) or t(17p), idic(X)(q13), t(1;3)(p36.3;q21.1), t(2;11)(p21;q23), inv(3)(q21q26.2), t(3;21)(q26.2;q22.1), t(6;9)(p23;q34), or t(11;16)(q23;p13.3).<sup>1</sup> These abnormalities are detectable by conventional cytogenetics and fluorescence in situ hybridization techniques. CGAT and other molecular techniques are essential for detecting other subtle abnormalities and cnLOH, which have proved to be an indicator of acquired disease. However, cnLOH is not included in the WHO’s list of recurring genetic abnormalities as evidence of MDS diagnosis.
In reporting CGAT results, our laboratory uses a filter size/resolution of 100 Kb for copy number gain and loss and 10 Mb for cnLOH abnormalities. We have reported cnLOH as a patient’s sole clonal abnormality suggesting disease; the frequency of this occurrence in the general patient population is unknown. We hope this case report will help broaden the awareness that the detection of cnLOH is important for early classification, treatment, and monitoring of MDS.
Patient case. We present a case of a 61-year-old male with a history of glioblastoma multiforme diagnosed in 2009 (Table 1). The patient was treated for glioblastoma with local radiation, tolerated an autologous transplant, received nine cycles of temozolomide, and achieved remission. The patient was followed with routine MRI but continued to experience fatigue. In November 2010 and March 2011, a full examination of the patient’s peripheral blood and bone marrow reported no abnormalities in myeloid blast, monocyte, or myeloid populations, or B or T cell populations. His results for conventional cytogenetics were consistently 46,XY[20] (no abnormalities) and normal FISH for chromosomes 5, 7, 8, 20, and the MLL locus at 11q23. CGAT was not performed at this time. During follow-up therapy, he continued to demonstrate extreme fatigue and his counts failed to rebound at a normal rate, which raised concern for aplastic anemia and treatment-related MDS. A brain MRI showed no evidence of tumor recurrence.
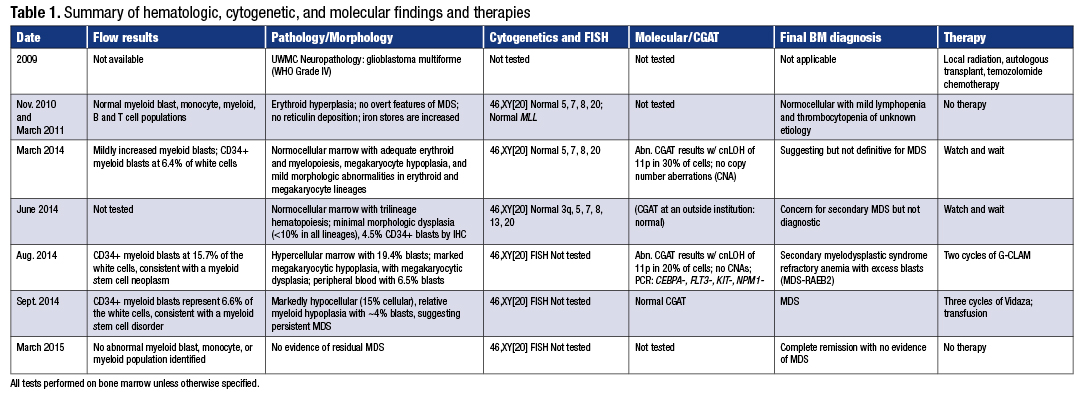 In March 2014 the patient requested evaluation at Seattle Cancer Care Alliance. A full examination of blood and bone marrow revealed concern for MDS with mildly increased myeloid blasts with mild immunophenotypic abnormalities and no evidence of glioblastoma multiforme (Fig. 1A and B, page 84). The CD34+ myeloid blasts represented 6.4 percent of the white cells by flow cytometry, and the abnormal cells by morphology were too low for the definitive diagnosis of MDS. The erythroid cells did show occasional irregular nuclear contours with megaloblastoid changes. The megakaryocytes were decreased in number with small hypolobulated forms. By morphology, the bone marrow blast count was three percent. The bone marrow biopsy showed a 30 percent cellularity. There were no ringed sideroblasts and no reticulin fibers. Flow cytometry showed a mildly increased myeloid blast population with mild immunophenotypic abnormalities. The cytogenetics and FISH continued to show normal results. However, results from the CGAT testing showed an abnormal result with clonal cnLOH of chromosome 11p of 38 Mb size in about 30 percent of cells (Fig. 2, page 86). No copy number aberrations were detected. Based on the lack of significant evidence of dysplasia by morphology, the patient’s disease did not meet the WHO-defined criteria for MDS diagnosis. However, the traits that were highly suggestive of MDS were the clinical setting of low blood counts following therapy with oral temozolomide, the mild dysplasia present, the immunophenotypic abnormalities observed by flow cytometry, and the clonal 11p cnLOH observed by CGAT. The providers decided to not initiate therapy but watch the patient’s progress closely.
In March 2014 the patient requested evaluation at Seattle Cancer Care Alliance. A full examination of blood and bone marrow revealed concern for MDS with mildly increased myeloid blasts with mild immunophenotypic abnormalities and no evidence of glioblastoma multiforme (Fig. 1A and B, page 84). The CD34+ myeloid blasts represented 6.4 percent of the white cells by flow cytometry, and the abnormal cells by morphology were too low for the definitive diagnosis of MDS. The erythroid cells did show occasional irregular nuclear contours with megaloblastoid changes. The megakaryocytes were decreased in number with small hypolobulated forms. By morphology, the bone marrow blast count was three percent. The bone marrow biopsy showed a 30 percent cellularity. There were no ringed sideroblasts and no reticulin fibers. Flow cytometry showed a mildly increased myeloid blast population with mild immunophenotypic abnormalities. The cytogenetics and FISH continued to show normal results. However, results from the CGAT testing showed an abnormal result with clonal cnLOH of chromosome 11p of 38 Mb size in about 30 percent of cells (Fig. 2, page 86). No copy number aberrations were detected. Based on the lack of significant evidence of dysplasia by morphology, the patient’s disease did not meet the WHO-defined criteria for MDS diagnosis. However, the traits that were highly suggestive of MDS were the clinical setting of low blood counts following therapy with oral temozolomide, the mild dysplasia present, the immunophenotypic abnormalities observed by flow cytometry, and the clonal 11p cnLOH observed by CGAT. The providers decided to not initiate therapy but watch the patient’s progress closely.
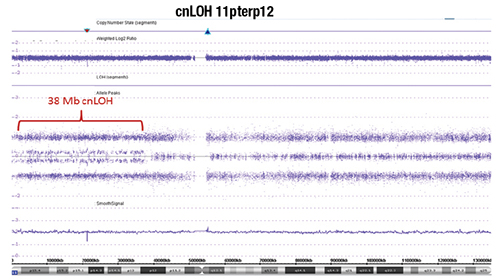
Fig. 2. Chromosome genomic array testing (CGAT) identified copy neutral loss of heterozygosity (cnLOH) of the short arm of chromosome 11 as the sole molecular abnormality. The X axis denotes genomic location (short arm 11p on the left and long arm 11q on the right separated by the centromere region with no probe coverage; see chromosome 11 ideogram on the bottom), while the Y axis denotes log2 ratio of the copy number (upper panel) and the allelic track (middle panel). Each blue/purple dot corresponds to a probe on the array. Chromosome 11q shows the normal allelic track pattern whereas 11p demonstrates cnLOH (splitting of the middle track) from the telomere (11pter) to band 11p12 (38 Mb in size) in approximately 30 percent of cells. There is no copy number aberration evident.
The patient was reassessed in August 2014. The flow cytometry revealed a significant increase of CD34-positive myeloid blast cells to 15.7 percent, consistent with a myeloid stem cell neoplasm. The morphology showed 19.4 percent blasts, marked megakaryocytic hypoplasia, with megakaryocytic dysplasia, consistent with MDS (Fig. 1C and D, page 84). The cytogenetic, FISH, and CGAT all showed results consistent with those reported in March; the cnLOH of 11p continued to be this patient’s sole detectable genomic abnormality while PCR results showed no mutations of CEBPA, FLT3, KIT, and NPM1 genes (Table 1). At this point the patient was classified as secondary MDS RAEB-2 and started G-CLAM chemotherapy. When the patient was evaluated in September 2014, his counts appeared to be recovering. By November 2014 he had no evidence of MDS and achieved complete remission. The patient’s platelet levels remained low and it was unclear if it was related to potential relapse of glioblastoma or minimal residual disease of MDS. Brain MRI in August 2015 confirmed brain tumor recurrence, and the patient died two months later without evidence of MDS.
Discussion. cnLOH was the only detectable abnormality in this patient’s molecular studies. CGAT is critical not only for the detection of cnLOH but also for submicroscopic genomic imbalances (copy number aberrations). These abnormalities are undetectable by conventional cytogenetics and FISH because they are below the threshold of detection size and resolution. In addition, because of a high degree of concordance with conventional cytogenetics and FISH, CGAT is effective at replacing imbalance FISH panels in the diagnostic setting.<sup>2</sup> In this case study, considering the lack of CNAs, chromosomal rearrangements, or common molecular aberrations detectable by PCR, the presence of cnLOH was the only genetic marker in which to follow this patient’s disease progression.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
