Sherrie Rice
March 2019—A small study performed at the University of Utah found that a next-generation sequencing assay cannot replace routine standard-of-care testing to detect pneumonia in immunocompromised patients and determine their treatment. But it could be ordered when an infection is suspected and all other testing has failed to find the etiology.
Brittany Young, MD, PhD, a fellow in medical microbiology in the university’s Department of Pathology, and colleagues set out to determine if one test, performed quickly and up front, could provide information on all organisms. Their study was funded by the pathology department and ARUP Laboratories.

Dr. Young
“Our current standard of care is to use bronchoscopy to collect lower respiratory tract specimens,” for which the published diagnostic yield is 31 to 80 percent, Dr. Young said last November in a presentation at the Association for Molecular Pathology annual meeting. “Routine standard-of-care testing uses large test panels, which include culture and targeted molecular testing and can take days to weeks to come to a resolution.”
Dr. Young’s colleagues at the University of Utah developed an unbiased metagenomics sequencing assay for bronchoalveolar lavage specimens, to be sequenced on an Illumina NextSeq 500. The turnaround time goal for the test in the clinical laboratory was 72 hours, “faster than a lot of routine testing,” she said, and potentially able to detect any virus, bacteria, or fungi present.
“Everything is analyzed with TaxonomerDx,” Dr. Young said, an online metagenomics analysis tool that analyzes RNA and DNA sequences but also maps them to a laboratory-developed reference database. “And it is done based on a Taxonomer score, which combines percent uniqueness, number of normalized reads, and depth of coverage. It can also exclude the human DNA.”
Dr. Young compared the results of their NGS assay with those of the university hospital’s standard test panel, known as immunocompromised hosts panel, or ICP, which includes a combination of culture, direct PCR, and targeted molecular testing. She also did a retrospective review of the charts for information about the patient’s clinical course, imaging results, medication lists, and more. “With this we wanted to look at two questions,” Dr. Young said. “Can NGS results correlate well with our routine standard of care but also detect additional possible pathogens negative by ICP, and what information can the NGS test provide to the clinician to improve patient care?” Their list of possible pathogens consisted of established respiratory pathogens and those that the clinical team deemed significant and treated.
The investigators studied 48 patients who had many types of immunosuppressive conditions and some of whom had bone marrow or solid organ transplants. Others had malignancies or autoimmune diseases that were treated with immunosuppressive agents. All had new radiographic evidence of infectious disease or consistent with such. Half ended up in the ICU and about half of them were intubated. Three died during hospitalization. “Although no autopsies were performed,” Dr. Young said, “the cause of death was thought to be attributed to respiratory failure in two of them. All but one of the patients were on antimicrobial therapy at the time of bronchoscopy, and this has been shown in previous studies to decrease diagnostic yield.”
The team found an overall positivity rate for the standard ICP of 62 percent. This NGS assay detected about 59 percent of the possible pathogens, and the overall agreement between the two assays was 52 percent. (See “Diagnostic yield of possible pathogens for ICP vs. NGS.”) Compared with the possible pathogens detected with the ICP, the NGS assay missed six of nine fungi, one of seven viruses, and four of nine bacteria. “These findings are the results from a study of respiratory specimens tested with our laboratory-developed NGS assay,” Dr. Young points out.
The team then studied the patient records to see if the NGS results would have altered treatment. They looked at 11 patients for whom only the laboratory’s standard-of-care testing detected the infections. “What happens potentially,” they wanted to know, “when NGS can miss these pathogens and the routine results are not available?” Dr. Young said.
One patient had methicillin-resistant Staphylococcus aureus. “The empiric therapy before BAL collection would not have covered MRSA,” she said. The patient was prescribed vancomycin once the ICP results were in. “If that organism had not been found, the patient might not have been on an effective therapy,” Dr. Young said.
Another patient had Enterobacter aerogenes. The culture susceptibility profile was used to determine that the patient needed to be switched to cefepime to cover that organism, similar to yet another patient who had Pseudomonas aeruginosa for whom the culture susceptibility panel allowed the clinicians to deescalate from the empiric cefepime to levofloxacin. “In another example, our patient with Coccidioides was on voriconazole for empiric treatment, but voriconazole is inferior to amphotericin B in this setting,” she said. “And five of our six patients who had Pneumocystis were missed by NGS, and half of these would not have been treated at all or would have been treated suboptimally.”
Empiric therapy covers most opportunistic pneumonias in this population and is generally effective, she said, but it is not a replacement for susceptibility testing provided by culture. NGS doesn’t sequence deeply enough to find all the drug resistance genes. Ten of their patients then would not have been treated optimally had only NGS been used.
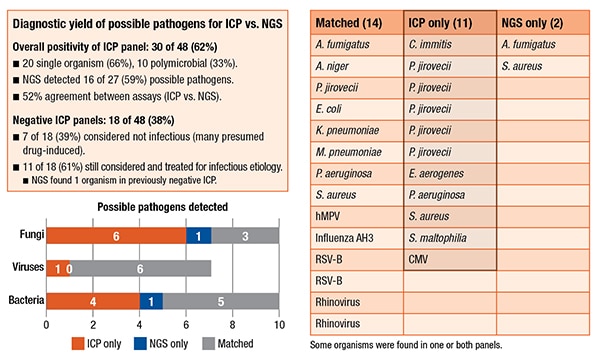 In other ways, however, their NGS assay did find more information than the ICP. There were 171 organisms in the 48 patients. Only 36 were true positives. (Taxonomer optimizes sensitivity over specificity, she pointed out, which creates a number of false-positives that have to be manually adjudicated using the Taxonomer score.) About 21 percent were adjudicated; 79 percent were not. The NGS assay was found to be slightly better at detecting viruses: 54 percent were adjudicated. It also detected two additional findings. In one patient who had a negative ICP, it found Aspergillus fumigatus. They also had a previously positive serum Galactomannan antigen test as well as a CT presentation that was “classic for Aspergillus. So the physicians were already treating the patient as an Aspergillus patient.” In another case, the NGS assay found an additional S. aureus and, in the culture, Enterobacter aerogenes grew, but there was also Gram-positive cocci on the Gram stain, so antibiotics would have covered that.
In other ways, however, their NGS assay did find more information than the ICP. There were 171 organisms in the 48 patients. Only 36 were true positives. (Taxonomer optimizes sensitivity over specificity, she pointed out, which creates a number of false-positives that have to be manually adjudicated using the Taxonomer score.) About 21 percent were adjudicated; 79 percent were not. The NGS assay was found to be slightly better at detecting viruses: 54 percent were adjudicated. It also detected two additional findings. In one patient who had a negative ICP, it found Aspergillus fumigatus. They also had a previously positive serum Galactomannan antigen test as well as a CT presentation that was “classic for Aspergillus. So the physicians were already treating the patient as an Aspergillus patient.” In another case, the NGS assay found an additional S. aureus and, in the culture, Enterobacter aerogenes grew, but there was also Gram-positive cocci on the Gram stain, so antibiotics would have covered that.
Their NGS assay also provides better resolution over the ICP. “We don’t normally differentiate our Candida species because they’re considered colonizers. But the NGS assay was able to detect and speciate Candida. It also specified some of the viral strains. So we think this may be useful for epidemiology studies,” Dr. Young said.
She listed the study’s limitations—small sample size, the use of frozen BAL specimens which may have affected analytical sensitivity, and the retrospective design—and provided the team’s main recommendation: The NGS assay on BAL specimens should be ordered only after all other testing has failed to find the etiology but an infection is still suspected. “The ideal of one test being performed at the beginning to catch everything,” she said, “is not yet ready.”
Sherrie Rice is editor of CAP TODAY. Dr. Young’s collaborators were Kimberly Hanson, MD, MPH, Daniel Thomas, and Taylor Snow.