Anne Ford
March 2018—When Stephen Brecher, PhD, compares MRSA to carbapenem-resistant Enterobacteriaceae, the figures of speech come fast and furious.
“We are not in Kansas anymore,” he said. “The bar has been raised. I consider MRSA a picnic compared to CP-CRE [carbapenemase-producing CRE].”
Dr. Brecher is director of microbiology and “chief of staph,” as he puts it, at the VA Boston Healthcare System and an associate professor of pathology and laboratory medicine, Boston University School of Medicine. Microbiology laboratories are the radar system needed to detect and prevent the spread of CP-CRE, Dr. Brecher says, and failure to do so would be catastrophic. There are concerns about using new, expensive tests, he acknowledges. But “the most expensive test is the one that does not work or takes too long to work.” Go to administration: “Plead your case to get the best technology available to do your job,” he says.
 Dr. Brecher spoke last fall on CP-CRE (carbapenemase-producing carbapenem-resistant Enterobacteriaceae), beta-lactamases, and antibiotic breakpoints in a webinar hosted by CAP TODAY and made possible by an educational grant from Cepheid. His co-presenter, Lars Westblade, PhD, of NewYork-Presbyterian/Weill Cornell Medical Center, spoke on the phenotypic and genotypic tests used to detect CP-CRE. (See page 16.)
Dr. Brecher spoke last fall on CP-CRE (carbapenemase-producing carbapenem-resistant Enterobacteriaceae), beta-lactamases, and antibiotic breakpoints in a webinar hosted by CAP TODAY and made possible by an educational grant from Cepheid. His co-presenter, Lars Westblade, PhD, of NewYork-Presbyterian/Weill Cornell Medical Center, spoke on the phenotypic and genotypic tests used to detect CP-CRE. (See page 16.)
Carbapenemases are enzymes that break down the structure of carbapenem antibiotics, of which there are four on the market: imipenem, meropenem, doripenem, and ertapenem. “We now have at least 500 different carbapenemases,” Dr. Brecher said, noting that resistance can be mediated by mechanisms other than through the carbapenemases.
Some places in the United States have had a lot of CP-CRE, but many places have seen very little. “Our goal has to be to keep it that way,” he said.
Consider this: With MRSA, there is one organism and one main resistance gene (mecA). It is relatively easy to grow Staphylococcus aureus and relatively easy to detect resistance, large segments of the population can be screened with a comparatively noninvasive anterior nares swab, and there are FDA-cleared chromogenic agars and nucleic acid amplification tests.
In contrast, “With CP-CRE, we have multiple organisms and multiple resistance genes. Although the organisms are usually easy to grow, it’s not always easy to detect resistance. When we do have carriers—and we don’t even know the actual carrier rate—detection is from a rectal swab, which is clearly more invasive than an anterior nares swab. There are no FDA-approved screening agars. There is an FDA-approved nucleic acid test” for about 90 of the most common of these genes in five main groups—KPC, NDM, VIM, OXA, and IMP. Antibiotic treatment can vary by class/type of carbapenemase.
The Centers for Disease Control and Prevention defines CP-CRE as Enterobacteriaceae that are resistant to imipenem, meropenem, doripenem, or ertapenem, or documentation that the isolate possesses a carbapenemase. “I respect the CDC but have a problem with their definition because it includes ertapenem,” Dr. Brecher said. “Ertapenem can be a marker for CP-CRE, but it has poor specificity and you’re going to get false-positives. That translates into time, money, and infection prevention dollars.”
The 2017 Veterans Affairs guidelines focused on E. coli, Klebsiella, and Enterobacter and left ertapenem and other Enterobacteriaceae out of the definition. “I am interested in other organisms as well, but just as thieves rob banks because that’s where the money is,” Dr. Brecher said, “our efforts really have to be at the big three.”

Dr. Brecher
Thus, Proteus, Providencia, and Morganella are not on the so-called hit list because they have natural, intrinsic low levels of imipenem resistance. Serratia and Citrobacter will be looked at more closely in the future. VA microbiology laboratories, and all others, are urged to use the most current CLSI breakpoints as well as rapid molecular confirmation. “I want to know to the best of my ability if the organism is a carbapenemase producer or if it has another mechanism of resistance, and that would also be used to guide treatment. Thankfully, there are a few new antibiotics that can be used to treat certain classes of CP-CRE.”
Just thinking about beta-lactamases can be overwhelming, Dr. Brecher said. “We have greater than 1,900 [of them].” They fall into four classes: molecular class A, which includes extended spectrum β-lactamases (ESBLs) and carbapenemases; molecular class B, which includes metallo-β-lactamases and metallo carbapenemases that require zinc; molecular class C, which includes the ampC SPICE and SPACE bacteria, but very rarely CRE; and molecular class D, some of which are weak carbapenemases with clinical importance.
For ESBLs—bacterial enzymes produced primarily by E. coli and Klebsiella species that break down the β-lactam ring of third- and fourth-generation cephalosporins—“the breakpoints were wrong,” Dr. Brecher said of his and others’ years-long work with the CLSI. “The ceftriaxone susceptible breakpoint used to be ≤ 8. For most patients with E. coli or Klebsiella, the MICs were nearly always ≤ 1. When the MICs were 2, 4, or 8, there were often treatment failures.
“The CLSI realized this important issue and worked on defining a methodology for detecting ESBLs,” he continued. “They came up with a very nice scheme. You tested selected third-generation cephalosporins with and without beta-lactamase inhibitors . . . and if you got a significant difference in the zone size, or a difference in the MIC, you could use that to say ‘This is an ESBL’ [or] ‘This is not an ESBL.’ And if it was an ESBL, the convention was to change all of the cephalosporins to resistant. That went on for a while but was clearly extra work for the lab.”
The CLSI solved the problem by changing the breakpoints.
“With the new, more stringent CLSI breakpoints, we could report the MIC, report the interpretation, and report those with elevated MICs as multidrug-resistant organisms,” he said.
The new CLSI breakpoints were also applicable for the class C β-lactamases—the ones that, for the most part, are chromosomal enzymes. “We knew that Enterobacter, Serratia, and Citrobacter constitutively made low levels of beta-lactamases.” Low enough that on initial testing they didn’t test resistant. “But if you got one from a patient’s bloodstream and put that patient on a beta-lactam antibiotic, the bacterial isolate could go from low-level production to high-level ampC beta-lactamase production, and within a few days the patient who had been doing well now crashes. It’s the same isolate as before but now it’s frankly resistant.”
More bad news: These chromosomal genes for ampC enzymes from Enterobacter, Serratia, and Citrobacter made their way onto plasmids, “and now we have plasmid-mediated transferable ampC genes in E. coli and Klebsiella.”
With CP-CRE, there are multiple classes. Within class A, “the biggest one, and the most important one, is Klebsiella pneumoniae carbapenemase,” he said. Class A KPCs are mostly found in K. pneumoniae and E. coli, but also in other Enterobacteriaceae. KPCbla genes reside in plasmids. Most KPCs hydrolyze all of the β-lactam antibiotics, with variable susceptibility patterns to other antibiotics. Twenty-four distinct types have been identified so far. They are endemic in New York City and have been found in other places in the United States. “They’re spreading, but as I said, they’re not everywhere in big numbers, and our job is to make sure it stays that way.”
Class B harbors plasmid-mediated metallo-β-lactamases (“metallo” indicates they require zinc for activity). The most common of them are the New Delhi metallo-β-lactamases, which have made it to the United States through medical tourism. “These have now spread to other organisms within the Enterobacteriaceae and even beyond the Enterobacteriaceae in organisms like Acinetobacter,” Dr. Brecher said.
He called the class D carbapenemases “very complicated.” They were originally described as OXA β-lactamases that could hydrolyze oxacillin and cloxacillin, but they also hydrolyze carbapenems. There are now five OXA families, with multiple enzymes in each family. They are primarily found in, but not limited to, Acinetobacter, Pseudomonas, and Enterobacteriaceae, and are not influenced by inhibitors such as EDTA or clavulanic acid. One very new, very mucoid K. pneumoniae strain, known as OXA-48, is problematic in many parts of the world, he said.
Dr. Brecher pointed out, too, that carbapenem resistance can be associated with other mechanisms. “If you have an isolate with an elevated MIC, it may be due to an ESBL along with a porin protein mutation.” This is more commonly seen for ertapenem. “Or you can have an ampC beta-lactamase and a porin protein or efflux mutation.”
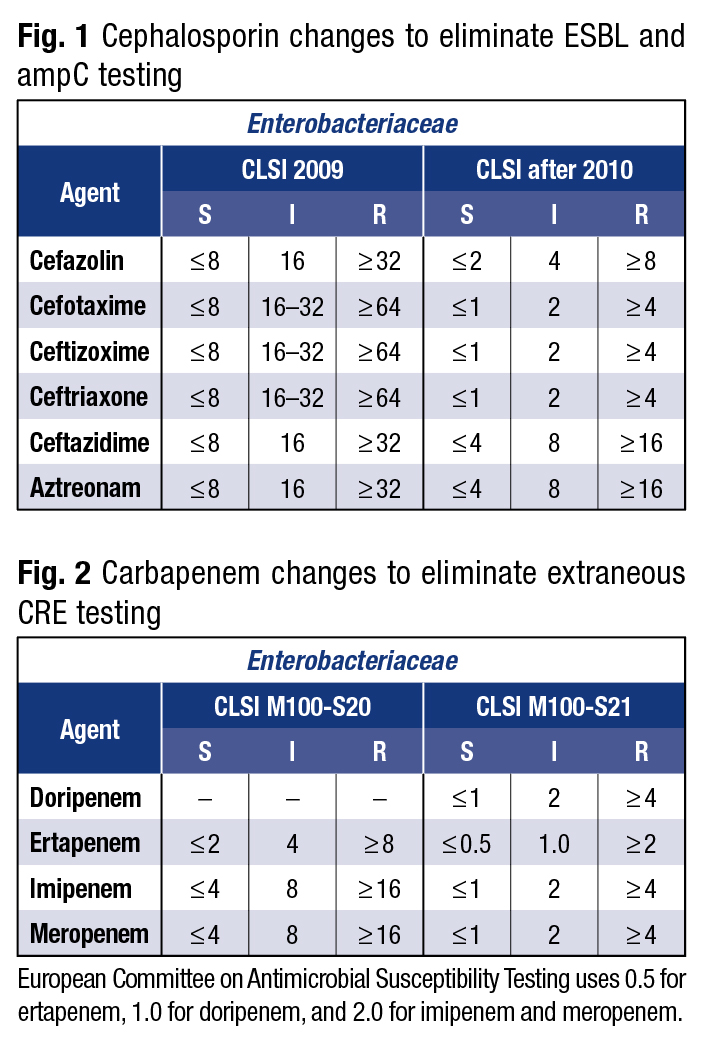
Breakpoints change because bacteria change, “and bacteria do not read or respect our rules,” Dr. Brecher said. The CLSI and European Committee on Antimicrobial Susceptibility Testing set the standards: What’s susceptible? What’s intermediate? What’s resistant? Laboratories must adapt to changed breakpoints to have a better chance of detecting CP-CRE. “I urge you to look at the breakpoints and change, so that you can do the best job that you possibly can,” he said. (See Figs. 1 and 2.)
As an example, he pointed to the fact that for years, 4 was considered susceptible for imipenem or meropenem—but then the breakpoint was changed to 1. “So if you get a carbapenemase producer that you confirm, let’s say by molecular technique, and it turns out that the MIC is 2 or 4, can you use that antibiotic?” Dr. Brecher asked. “And the answer may be yes; it will depend on the organism and the site of the infection.” Microbiologists, pharmacists, and physicians have to communicate. “MICs are complicated and antibiotic specific. Communication is key.”
Several new antibiotics geared toward CP-CRE are now available, such as the FDA-approved ceftazidime + avibactam (Avycaz) and meropenem + vaborbactam (Vabomere). “That’s really good news,” Dr. Brecher said. At the same time, there has been a recent report of the isolation of E. coli and K. pneumoniae from animals and humans in China harboring plasmids for mobilized colistin resistance (mcr-1) and for a metallo carbapenemase (NDM-5), as well as evidence that these strains have spread to South Asia, Europe, South Africa, and South America, and have spread within Enterobacteriaceae. “It is tough trying to stay ahead of evolving resistance in bacteria,” he said.
“Think of it: Bacteria are 3.8 billion years old and we have been using antibiotics for 75 years. Time to roll up our sleeves.”
[hr]
Anne Ford is a writer in Evanston, Ill The full webinar is available at www.captodayonline.com.