Amy Carpenter Aquino
March 2023—Assessing cardiac allograft rejection from endomyocardial biopsy and assigning a differential diagnosis to cancers of unknown origin have been shown to get a boost from AI-driven computational pathology models. So too has identifying subregions of high diagnostic value on whole slide images.
In addition, an algorithm published late last year addresses familiar challenges in whole slide image search: speed, accuracy, and scalability.
Faisal Mahmood, PhD, spotlighted these computational pathology models when he spoke at the Association for Molecular Pathology meeting last November. Dr. Mahmood, associate professor in the Department of Pathology at Harvard Medical School, and in the Division of Computational Pathology at Brigham and Women’s and Massachusetts General hospitals, shed light on what the field of computational pathology is aiming now to do.
His group studies phenotypic and morphologic data and performs quantitative spatial analysis to provide early diagnoses and prognoses, predict response to treatment, stratify patients, and discover biomarkers. They also look at integrative sources that are already quantitated, such as multimodal or molecular biomarkers, and use deep learning to integrate the information. “We’re also interested in genotypic and phenotypic responses to disease,” says Dr. Mahmood, who is also a member of the Cancer Data Science Program at Dana-Farber Cancer Institute and of the Cancer Program at Broad Institute of Harvard and MIT.
Conventional algorithms are difficult to use with typically large pathology images, he noted. As departments move to digital pathology-based workflows, “our hope is we will have a wealth of data” and “be able to do things we were never able to do before—rediscover diseases we already knew about and discover new ones, discover new morphologic features, correlate the morphology with molecular information in a more holistic manner.”
While supervised learning methods require manual labeling or region of interest extraction, deep learning does not require feature engineering, “so you don’t need to use existing human knowledge to handcraft very fine details within the image,” Dr. Mahmood said. “Annotations are enough.”
But annotating whole slide images is time-consuming and “far apart from what a clinical workflow looks like,” he said. The solution: weakly supervised learning, which requires only information that is already on WSIs and slide-level labels from pathology reports. Graph computational networks and multiple-instance learning are two common approaches.
Dr. Mahmood and colleagues in 2021 reported on a weakly supervised deep-learning method for data-efficient whole slide image processing and learning that requires only slide-level labels (Lu MY, et al. Nat Biomed Eng. 2021;5[6]:555–570). Their AI-driven method, called clustering-constrained-attention multiple-instance learning (CLAM), “uses a number of different bells and whistles from conventional machine learning and brings them together and applies them to pathology images,” he said. CLAM uses attention-based learning to identify the most important subregions within a WSI—to accurately classify the whole slide—and uses “tricks,” he said, to deal with data efficiency and imbalances. It also makes use of powerful pretrained ResNet encoders: “We use features learned on real-world images and then apply them to pathology images,” Dr. Mahmood explained.
The CLAM method segments and patches a WSI into smaller patches. “Once we have smaller patches, we extract features in a lower dimensional representation,” he said. An initial study used the conventional ResNet-50 encoder pretrained on real-world images, using just the first few blocks. “From there we learned more common, basic features of an image.” The intention, he said, was to learn what the most important regions are within the WSI. The model used N parallel attention branches that calculate N unique slide-level representations to enable multiclass classification. “Pathology in general is multiclass, and there could be many, many findings” in a WSI, he said.
“Then we rank the patches,” Dr. Mahmood said, and the patches are then pooled based on their learned rank to get to the slide-level representation. “The trick we use is instance-level clustering”—clustering similar morphologic images within the WSI. “Because we have attention built in and can rank the patches, we can project that back onto the whole slide image as an interpretability mechanism to make it a little more transparent than what features the model uses in making these classification determinations,” he said.
In the initial study, his group applied the model to renal cell cancer subtyping, non-small cell lung cancer subtyping, and detection of breast cancer lymph node metastasis. For the breast cancer lymph node metastasis, the WSI—“we used about 1,000 images”—was adapted to an independent cohort from Brigham and Women’s Hospital. He and his coauthors reported strong performance and the ability to generalize to independent test cohorts and varying tissue content. They write, “Our analysis demonstrated that CLAM can be used to train interpretable, high-performance deep-learning models for both binary and multi-class WSI classification using only slide-level labels without any additional annotation.”
Said Dr. Mahmood, “As we go from using about 700 slides to train this all the way down to 100 slides, there’s a drop in performance, but the drop in performance is not that large for this particular method, which means this approach, by using whole slide images, just labels that are available in pathology reports, and data efficiency, can be used for a lot of downstream tasks.”
To test how far they could push the model’s adaptability, Dr. Mahmood’s group tested CLAM with images taken with a cell phone connected to a brightfield microscope. “We found it was possible,” though there was an expected drop in performance in non-small cell lung cancer subtyping and renal cell carcinoma subtyping.
“So now that we had a mechanism to train these models on whole slide images using slide-level labels and doing it in an easy, effective manner, we planned to target the problem of cancer of unknown primary origin,” Dr. Mahmood said.
He and colleagues questioned whether it was possible to use conventional WSIs to predict tumor origin. They trained their new deep-learning–based algorithm using 22,833 WSIs from cases with known primary origin from The Cancer Genome Atlas and Brigham and Women’s Hospital, and from MGH in subsequent versions. The deep-learning algorithm—tumor origin assessment via deep learning (TOAD)—was then tested on an internal cohort of 6,499 primary and metastatic cases, Dr. Mahmood said. They had an external test cohort of 682 cases from 223 medical centers. And they assessed their model using an additional test data set of 317 cases (from 152 cen-ters) of cancer of unknown primary that were assigned a primary differential based on ancillary tests, radiology, history, clinical correlation, or at autopsy (Lu MY, et al. Nature. 2021;594[7861]:106–110).
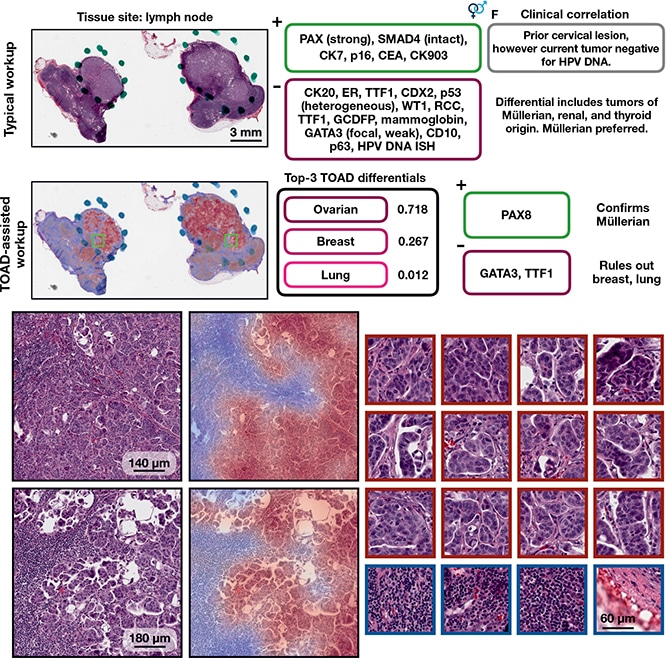
TOAD-assisted CUP workup: example 1. Top, a representative case that underwent a standard CUP workup involving extensive IHC staining and clinical correlation. Strong PAX8 staining suggested a Müllerian origin and multiple IHC tests were used to rule out other primary tumors. Retrospectively, they analyzed the case with TOAD and found that the top-3 determinations were ovarian, breast, and lung, and, after this determination, that only three IHC stains (PAX8, GATA3, and TTF1) needed to be used to confirm a Müllerian origin and rule out breast carcinoma and lung adenocarcinoma. This workflow demonstrates how TOAD can be used as an assistive diagnostic tool. Bottom, medium magnification and corresponding heat maps of representative areas of tumor, with high-magnification, high-attention patches on the right outlined in crimson and low-attention patches outlined in navy. Ming Y. Lu et al. AI-based pathology predicts origins for cancers of unknown primary. Nature. 2021;594(7861):106–110, Springer Nature.
The TOAD architecture was similar to the CLAM architecture but it was posed as a multitask problem, Dr. Mahmood said. “The model can predict whether the case is primary or metastatic and assign possible origins” for use in diagnosis, research, and guiding downstream ancillary testing. And “it did quite well on both internal and external test sets,” he said. “Wherever the model was more confident, it would make more accurate predictions.” On the test set of 6,499 (not seen by the model during training), it achieved an overall accuracy of 83.4 percent. He and his coauthors write, “When the model is evaluated using top-k differential diagnosis accuracy—that is, how often the ground truth label is found in the k highest confidence predictions of the model—TOAD achieved a top-3 accuracy of 95.5% and top-5 accuracy of 98.1%.”
“The top-three and top-five predictions were very high,” Dr. Mahmood said, “which means the model could be used to tell what the possible origins are and subsequently to order IHC tests or other ancillary tests” based on that information.
With limited time to follow all patient outcomes, the study authors divided the 317 cases with a primary differential into 193 cases with high certainty differential and 124 cases with low certainty differential and found 61 percent of cases agreed with the model’s prediction. The top-three agreement was 82 percent; the top-five agreement was 92 percent. “The top-three and top-five predictions were still very, very high,” Dr. Mahmood said.
More recently, his group has been trying to integrate more information for origin prediction. “We’ve found in general that integrating both histology and genomics has improved the prediction of origins across the board within the bounds of the study,” he said, noting they’re working now on expanding it. “We’re also able to explicitly go in and look at what morphologic origins are used in the whole slide image and then what’s used and what’s important within the molecular profile for each one of the origins we have used.”
Published last year was their report on their weakly supervised deep-learning approach for cardiac allograft rejection screening in H&E-stained WSIs, called cardiac rejection assessment neural estimator (CRANE) (Lipkova J, et al. Nat Med. 2022;28[3]:575–582). The model architecture is similar to the earlier model and adapted to the inter- and intraobserver interpretability problem in endomyocardial biopsy.
“We used about 1,300 cases from the Brigham and Women’s Hospital to develop the model,” Dr. Mahmood said, and adapted it to internal cases and to cases sent to them from hospitals in Turkey and Switzerland.
“We used different data collection mechanisms by deliberately using different slide scanners,” he said, and the staining protocols across the international cohorts differed as well. CRANE performed well on internal cohorts, but there was an expected drop in performance when adapting the model to the Turkish and Swiss data. He and coauthors write, “The model detects allograft rejection with an AUC of 0.962, assesses the cellular and antibody-mediated rejection type with AUCs of 0.958 and 0.874, respectively, detects Quilty-B lesions, benign mimics of rejection, with an AUC of 0.939, and differentiates between low- and high-grade rejections with an AUC of 0.833.”
This model, they write, “demonstrates the promise of AI integration into the diagnostic workflow,” though “optimal use of weakly-supervised models in clinical practice remains to be determined.”
In another study published in 2022, Dr. Mahmood and colleagues used weakly supervised multimodal deep learning to examine pathology WSIs and molecular profile data from 14 cancer types (Chen RJ, et al. Cancer Cell. 2022;40[8]:865–878). Their algorithm was able to “fuse these heterogeneous modalities to predict outcomes and discover prognostic features that correlate with poor and favorable outcomes,” the authors write. WSIs can be used to solve patient ranking problems, Dr. Mahmood said. “In this particular case we use overall survival.” Computational models developed using WSIs reported on in their earlier studies used just histology and WSIs to go directly to survival and other outcome predictions. “In this case, we’re integrating molecular information as well” in a limited setting using WSIs from The Cancer Genome Atlas.
“So this could result in better prognostic models, but perhaps the more interesting aspect here is that we can go in and look at what was important in the morphologic profile, what was important in the molecular profile, and how these things shift when additional modalities are included,” he said.
He and his coauthors found they can separate high-risk versus low-risk distinctions in 10 of the 14 cancer types they studied. The more interesting result, he said, is the analysis showing which cancer types would benefit from algorithms built using only WSIs, which cancer types would benefit from using molecular information alone, and for which ones it would be beneficial to include histology and molecular information in making prognostic determinations.
WSIs on average accounted for 16.8 percent of input attributions in multimodal fusion for all cancer types, which the authors say “suggests that molecular features drive most of the risk prediction” in multimodal fusion. However, for multimodal fusion models evaluated on uterine corpus endometrial carcinoma, “WSIs contributed to 55.1% of all input attributions,” the authors report. They also observed relatively larger average WSI contributions in head and neck squamous cell carcinoma, liver hepatocellular carcinoma, and stomach adenocarcinoma.
“But also on the disease level, we can quantify our architecture looking at the whole slide level heat maps and associate the two,” Dr. Mahmood said. Quantitative determinations are made for the features used in high-risk versus low-risk patients. For example, higher lymphocyte numbers are seen in low-risk patients, and lower lymphocyte numbers in high-risk patients, he said. “It’s a similar analysis in all 14 cancer types.”
More recently, they have incorporated radiology information into building their prognostic models and risk profiles. “In general,” he said, “we’re able to show that for a variety of disease models, we’re able to separate the patients very well into distinct groups and do better risk stratification if we use multiple modalities and data types.”
The group most recently reported on its self-supervised image search for histology algorithm, or SISH (Chen C, et al. Nat Biomed Eng. 2022;6[12]:1420–1434). Retrieval speeds of algorithms for searching similar WSIs often scale with repository size, which limits their clinical and research potential, Dr. Mahmood and coauthors write, and in this study they show that self-supervised deep learning “can be leveraged to search for and retrieve WSIs at speeds that are independent of repository size.” Image retrieval or retrieval of similar cases or just an image search is particularly important for rare diseases for which the number of available WSIs is often too low to train supervised deep-learning models, Dr. Mahmood noted. In developing the SISH model, “we tried to address a number of different issues with post-slide image retrieval.”
From each patch in the mosaic from a WSI, “we extract features using an encoder trained on self-supervised learning and another encoder trained on conventional images,” he said. “And the self-supervised encoder in this case was trained with a discrete latent code,” though other self-supervised encoders could be used.
WSI retrievals are “notoriously memory-hungry, but we found that for this particular problem we were able to use a consumer-grade workstation,” Dr. Mahmood said.
The algorithm was tested on cases from Brigham and Women’s and Massachusetts General and the TCGA, and “in general, over very extensive testing across 22,000 cases, it worked quite well,” he said. Their analysis showed how the model scales with increasing amounts of data. The speed “stays almost constant in searching for similar cases, which could be quite important if this were to be used in a diagnostic setting.” His group is working to deploy SISH at MGH.
The authors write: “Our experiments demonstrate that SISH is an interpretable histology image search pipeline that achieves constant search speed after training with only slide-level labels. We also demonstrate that SISH has strong performance on large and diverse datasets, can generalize to independent cohorts as well as rare diseases and, finally, that it can be used as a search engine not just for WSIs, but also for image patch retrieval.”
While the computational pathology “story so far” seems to have solved a lot of problems, Dr. Mahmood said, challenges remain. The chief limitation is reduced communication between smaller patches once they are patched. “The models are often not very context-aware,” he said, which is “a major problem in computational pathology.” In addition, few samples are available.
In natural language processing, the context matters a lot. In whole slide images, each resolution level can be seen to convey a different story: “Cellular features lead to slide-level organization, phenotypes to slide-level diagnosis, essentially. And we wanted to see if we could use methods commonly used in natural language processing and build a newer architecture using self-supervised learning to cater to some of the context-aware issues,” Dr. Mahmood said.
“So that’s exactly what we did.” Dr. Mahmood’s group’s submission to the 2022 IEEE/CVF Computer Vision and Pattern Recognition Conference described how to use whole slide images and patch them, going to patch-level and cellular-level representations. The result is a hierarchical image pyramid transformer architecture with patch-level, region-level, and slide-level representations.
“We’ve shown this essentially leads to much better results,” Dr. Mahmood said. The model was trained on TCGA cases, “and now we’re trying to scale it to a much more generic, much larger data set, hoping to convert it to a model that can be used for a variety of downstream tasks.”
Amy Carpenter Aquino is CAP TODAY senior editor.