November 2018—Color Atlas of Mycology: An Illustrated Field Guide Based on Proficiency Testing is a new book from CAP Press, released in October. It is designed to help in identifying fungi using the most recent taxonomic classifications. In it is more than 15 years of proficiency testing data to highlight diagnostic clusters of incorrect identifications and address conceptual classification issues. Following is an excerpt from the section on yeast.
Authors of the new atlas are Gordon L. Love, MD, chair of the Department of Pathology, Louisiana State University School of Medicine, and director of laboratories, University Medical Center, New Orleans; and Julie A. Ribes, MD, PhD, a professor of pathology and laboratory medicine and the director of the clinical microbiology laboratory and assistant director of special chemistry, infectious disease serology, University of Kentucky Medical Center, Lexington.
Essential Facts
 Culture Characteristics: Candida famata grows rapidly on routine mycology media to produce cream-colored colonies at 25°C. It produces oval budding yeast on cornmeal Tween-80 agar that are about 3.5 µm to 5 µm in diameter. Mycelia are usually absent, but rudimentary pseudohyphae may occasionally form.
Culture Characteristics: Candida famata grows rapidly on routine mycology media to produce cream-colored colonies at 25°C. It produces oval budding yeast on cornmeal Tween-80 agar that are about 3.5 µm to 5 µm in diameter. Mycelia are usually absent, but rudimentary pseudohyphae may occasionally form.
Potential Look-Alikes: By cornmeal Tween-80 morphology, Candida famata, C glabrata, C inconspicua, and Saccharomyces cerevisiae also produce yeast without pseudohyphae. C famata is prone to misidentification by automated biochemical microbial identification systems.
Key Differentiating Feature: Candida famata identification by molecular means or MALDI-TOF may be helpful.
Clinical: Candida famata is an unusual human pathogen, with few bloodstream infections reported.
Ecology: Candida famata is commonly isolated from dairy products.
MALDI-TOF: An accurate identification of this yeast is possible using either FDA-approved Bruker Biotyper CA or VITEK MS PLUS system. Both systems currently list this organism as Candida famata.
Colonial Morphology
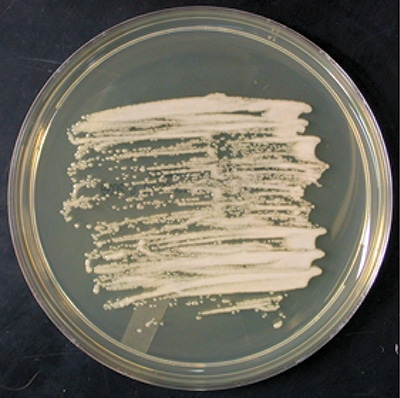
Figure 14-1. Candida famata grows rapidly on routine mycology media to produce cream-colored colonies at 25°C. Growth is variable on media containing cycloheximide. Maximum growth temperature is 35°C to 37°C.1
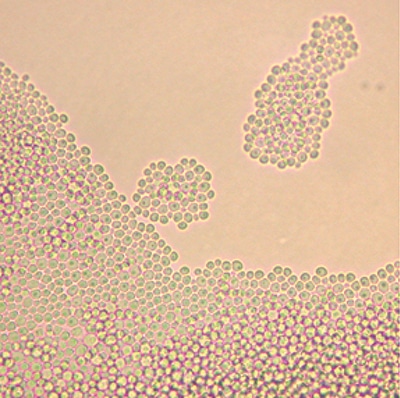
Figure 14-2. Candida famata produces oval budding yeast on cornmeal Tween-80 agar that are about 3.5 µm to 5 µm in diameter.2 Mycelia are usually absent, but rudimentary pseudohyphae may occasionally form (cornmeal Tween-80 agar, original magnification ×400).
Microscopic Morphology
Conjugation between a mother and its bud or between separate cells may produce sexual ascospores. The sexual forms or asci retain the same sizes as the yeast of 3.5 µm to 5 µm. Ascospores are spherical forms that have been described as warty, but the warty appearance is not well distinguishable under light microscopy. One or two ascospores develop per ascus. Ascospore development has been observed on V8, Gorodkowa, McClary’s acetate, and YM agars after 1 to 2 weeks of incubation at 15°C to 20°C.3
Candida famata has a physiological profile very similar to Candida guilliermondii complex, C intermedia, and Wickerhamomyces anomalus (formerly C pelliculosa).1 As a result, identification of C famata by commercial yeast identification panels and systems may be limited. MALDI-TOF and molecular sequencing are preferable for definitive identification.4 Approximately one-half of C guilliermondii complex isolates produce short pseudohyphae in addition to oval yeast—a feature that would help discriminate the yeast from C famata.
Taxonomy
Candida famata was formerly known as Torulopsis candida. After the Amsterdam Declaration on Fungal Nomenclature in 2013, Debaryomyces hansenii, the name of the teleomorph (sexual form), is preferred over the anamorph (asexual) name of C famata, although adoption of this new nomenclature is not readily occurring. FDA-approved identification systems still list the organism as Candida famata.
By phylogenetic analysis, Candida famata is closely related to C guilliermondii.
Proficiency Testing
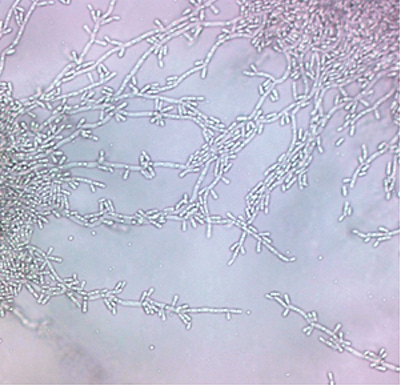
Figure 14-3. Candida zeylanoides (above) produces elongated blastoconidia at junctions between pseudohyphae. Prominent pseudohyphae should not be seen with Candida famata (cornmeal Tween-80 agar, original magnification ×400).
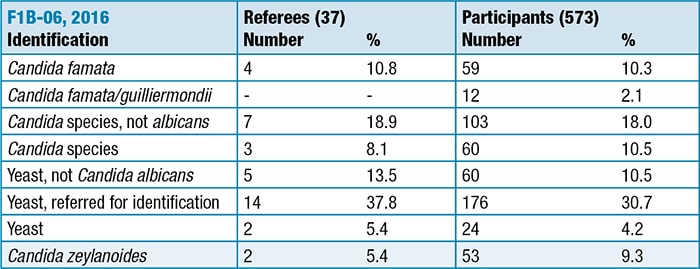
Only 59 laboratories (10.3%) identified Candida famata to species. An additional 12 laboratories (2.1%) selected the combined Candida famata/guilliermondii category. A significant distractor was C zeylanoides (Figure 14-3) by 53 laboratories (9%). These laboratories reported that biochemical identification systems produced the erroneous identification, which is unexpected in that C zeylanoides will not ferment carbohydrates and only assimilates glucose. In contrast, C famata has a very active assimilation and fermentation profile.
Few participants chose a combined category of Candida famata/C guilliermondii in the 2016 CAP Master List of Fungi. The suggestion that C guilliermondii is a likely misdiagnosis of C famata appears to be false.
Disease
In a study of 26 environmental and clinical isolates identified as Candida famata, only three were confirmed as C famata, six were identified as C caribbica, 10 were identified as C guilliermondii, two were C haemulonii, two were C lusitaniae, and three were C palmioleophila.5 In a more recent study of 53 C famata isolates collected during SENTRY and ARTEMIS surveillance programs that had been identified by a variety of commercial methods, DNA sequencing found that 19 were C guilliermondii, 14 were C parapsilosis complex, five were C lusitaniae, four were C albicans, three were C tropicalis, five were other Candida species, and three were non-Candida yeast.6 None were C famata. The ability to identify C famata is limited except for MALDI-TOF MS or DNA sequencing.
 Poor discrimination for Candida famata among manual and automated fungal identification systems contributes to the scarcity of C famata in reports of disease. C famata infections appear to be related to previous surgery, indwelling catheters, and immunosuppression.7
Poor discrimination for Candida famata among manual and automated fungal identification systems contributes to the scarcity of C famata in reports of disease. C famata infections appear to be related to previous surgery, indwelling catheters, and immunosuppression.7
References
- de Hoog GS, Guarro J, Gené J, Figueras MJ. Debaryomyces hansenii (Zopf) Lodder & Kreger-van Rij. Atlas of Clinical Fungi: The Ultimate Benchtool for Diagnostics. Version 4.1.2. CBS Fungal Biodiversity Centre, Utrecht, The Netherlands. www.clinicalfungi.org. July 2014. Accessed August 24, 2016.
- St-Germain G, Summerbell R. Identifying Fungi: A Clinical Laboratory Handbook. 2nd ed. Belmont, CA: Star Publishing Co; 2011:94–95.
- Kurtzman C, Fell JW, Boekhout T, eds. The Yeasts: A Taxonomic Study. 5th ed. London, England: Elsevier; 2011:364–365.
- Westblade LF, Jennemann R, Branda JA, et al. Multicenter study evaluating the VITEK MS system for identification of medically important yeasts. J Clin Microbiol. 2013;51(7):2267–2272.
- Desnos-Ollivier M, Ragon M, Robert V. Debaryomyces hansenii (Candida famata), a rare human pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J Clin Microbiol. 2008;46(10):3237–3242.
- Castanheira M, Woosley LN, Diekema DJ. Candida guilliermondii and other species of candida misidentified as Candida famata: assessment by VITEK 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry in two global antifungal surveillance programs. J Clin Microbiol. 2013;51(1):117–124.
- Bayda ND, Chuang SH, Alam MJ, et al. Treatment of Candida famata bloodstream infections: case series and review of the literature. J Antimicrob Chemother. 2013;68(2):438–443.
To order, go to cap.org, “Shop” tab, or call the CAP at 800-323-4040 option 1 (PUB226). CAP member price: $144; for others: $180. Ebook: $130 at ebooks.cap.org.