Jonas J. Heymann, MD; Mahesh M. Mansukhani, MD; Anthony N. Sireci, MD; Hui-Min Yang, MD; Pokala R. Kiran, MD; Antonia R. Sepulveda, MD, PhD
August 2017—CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from Columbia University Medical Center. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Collision tumors are thought to represent the chance encounter and temporospatial interaction of independent tumors, and their accurate classification may have significant prognostic and therapeutic implications. Here we showcase a next-generation sequencing panel used to analyze both components of a potential collision tumor and render the correct diagnosis.
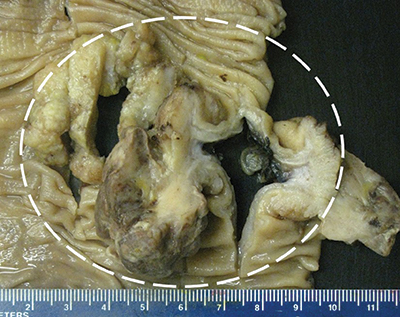
Fig. 1. (A) Ileocolectomy specimen with grossly identifiable fungating mass—outlined in white—extending into pericolic adipose tissue and centrally located firm, white area.
Case presentation and history. The patient was an 87-year-old woman and former smoker who presented with hematochezia and dyspnea on exertion. She denied a history of gynecologic malignancy, uterine fibroids, or vaginal bleeding. Computed tomography of the abdomen and pelvis demonstrated a 6-cm right colonic mass without mesenteric lymphadenopathy. She was found to be severely anemic, with a plasma hemoglobin level of 5.5 g/dL, and to have serum carcinoembryonic antigen level elevated at 46.7 ng/mL. Diagnostic colonoscopy was aborted after the patient developed atrial fibrillation with a rapid ventricular response. Therefore, the patient underwent ileocolectomy.
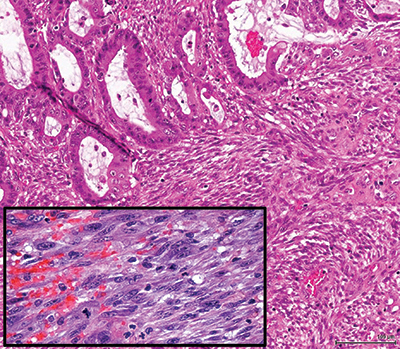
Fig. 1. (B) Microscopically, an adenocarcinoma interdigitates with a malignant spindle cell proliferation. Photomicrograph was taken at 200× (inset, 400×).
Results. Gross examination showed a single 6.6-cm fungating mass extending through the muscularis propria into pericolic adipose tissue. Sectioning revealed an ill-defined firm, white area measuring approximately 3 cm. Histologically, the tumor was a composite of two interdigitating patterns. First, an adenocarcinoma with well-to-moderate differentiation and mucinous features arose from an adenoma with focal high-grade dysplasia. Immunohistochemical analysis demonstrated that adenocarcinoma cells expressed pan-cytokeratin (CK), CK20, and caudal-related homeobox protein-2 (CDX2) but not CK7. Second, the firm, white area identified grossly consisted of a spindle cell proliferation with fascicular architecture, focal necrosis, and high mitotic rate (up to 17 mitoses in 10 high-power fields). IHC analysis demonstrated that malignant spindle cells expressed desmin and smooth muscle actin but not pan-CK, CK7, CK20, CDX2, or markers of gastrointestinal stromal tumor, gynecologic malignancy, and neural and mesothelial origin, suggestive of smooth muscle differentiation. The differential diagnoses included carcinosarcoma and collision of adenocarcinoma and leiomyosarcoma. The carcinoma and sarcoma were staged separately as pT3N0 and pT1bN0, respectively.
IHC analysis also demonstrated loss of expression of MLH1 and PMS2 in the adenocarcinoma component, consistent with a DNA mismatch repair-deficient tumor, whereas expression of all mismatch repair proteins (MLH1, MSH2, MSH6, and PMS2) was preserved in the sarcomatous component. Accordingly, microsatellite instability testing demonstrated high-frequency MSI in the adenocarcinoma but not in the sarcoma.
Mutational analysis was performed separately on the two tumor components by next-generation sequencing with a cancer gene panel. The area of carcinoma demonstrated the following mutations in cancer genes:
- c.1799T>A missense mutation (NM_004333.4, p.Val600Glu) in the BRAF gene with 22 percent variant allelic fraction;
- c.451delC frameshift deletion (NM_000546.4, p.Pro151fs) in the TP53 gene with 22 percent allelic fraction;
- c.3062A>G missense mutation (NM_006218.2, p.Tyr1021Cys) in the PIK3CA gene with 24 percent allelic fraction;
- c.1624delA frameshift deletion (NM_000368, p.Lys542fs) in the TSC1 gene with 15 percent allelic fraction; and
- 80 nucleotide variants of uncertain significance.
The sarcomatous component demonstrated a c.2629_2647del frameshift deletion (NM_001042492.2, p.Met877fs) in the NF1 gene with 19 percent allelic fraction, as well as six variants of uncertain significance.
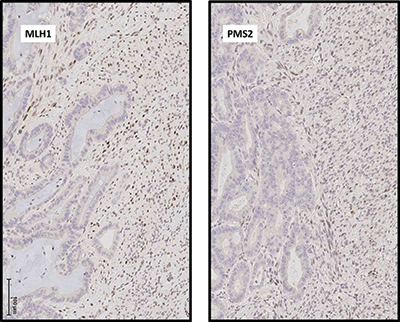
Fig. 2. (A) Immunohistochemical analysis demonstrates that expression of DNA mismatch repair proteins MLH1 and PMS2 is absent in malignant glandular epithelial cell nuclei but intact in malignant spindle cells. Photomicrographs taken at 200×.
Discussion. Leiomyosarcoma of the gastrointestinal tract is a rare tumor, and the finding of adenocarcinoma closely associated with a high-grade spindle cell proliferation raises two main differential diagnoses: carcinosarcoma, either primary or metastatic from a site such as the gynecologic tract, and collision of mucinous adenocarcinoma and leiomyosarcoma of the colon. The combined evaluation of IHC patterns and molecular findings in our case include distinct MSI status of the two separate tumor components and distinct mutational profiles. It is likely that these lesions originate from different clones and via different molecular pathways, and thus represent a collision tumor rather than carcinosarcoma.
This case represents the most extensive genetic analysis of a collision tumor of the colon yet reported, and it is the first report of NGS employed to analyze a collision tumor with carcinomatous and sarcomatous components. A previous report of a colorectal collision tumor included loss of heterozygosity in two of three microsatellite markers in a metastatic gastric carcinoma colliding with a primary rectal carcinoma with retention of all three markers. Both components of the collision tumor demonstrated microsatellite stability.1 A previous analysis of a thoracic collision tumor with pulmonary adenocarcinoma and malignant mesothelioma components included copy number analysis by single nucleotide polymorphism microarray and mutational analysis on a limited, amplicon-based NGS platform. Despite significant differences in genomic regional copy number, the NGS panel was too limited to identify a driver mutation in either tumor.2 The more extensive NGS panel used in our case allowed for identification of driver mutations in both tumors. Despite substantial evidence of mismatch repair deficiency in the adenocarcinoma, no loss-of-function mutation was identified in any mismatch repair gene. In such cases, mismatch repair deficiency may be caused by MLH1 gene promoter hypermethylation. Although the MLH1 promoter was not specifically interrogated in the current case, a tight association between activating BRAF mutation and MLH1 promoter hypermethylation is well established.3,4 As an activating mutation in BRAF was identified by NGS in the current case, MLH1 promoter hypermethylation is the likely cause of mismatch repair in the current case.
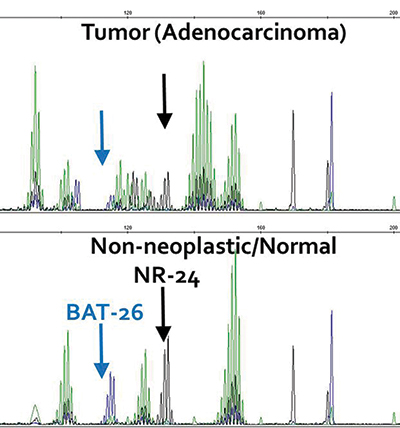
Fig. 2. (B) Microsatellite testing of the adenocarcinoma demonstrated instability of 5/5 mononucleotide repeats, two of which are highlighted here.
Therapy for collision tumors may need to be individualized for each tumor component. In this case, where neither component showed evidence of metastatic disease, surgical therapy alone was curative. Follow-up at one year after surgery did not show evidence of recurrence or metastatic disease. Thus, no further therapy was required.
In clinical practice NGS is used to provide predominantly predictive and prognostic information in most cancer cases. In this case, should the carcinoma metastasize, its MSI status would make the patient potentially eligible for immune checkpoint inhibition. Due to the presence of BRAF V600E mutation, the patient would be potentially eligible for trials of BRAF inhibitors and alternative chemotherapy combinations.5 Should the sarcoma recur or metastasize, the presence of an inactivating NF1 mutation could make the patient eligible for trials of MEK inhibitors and other therapies. However, in this case of a possible collision tumor, it is especially important to note that extended gene panel sequencing provided diagnostic information, in addition to predictive information, complementing morphologic and immunohistochemical analysis.
Methods. MSI testing was performed using the Promega (Madison, Wis.) MSI system according to the manufacturer’s instructions. Briefly, a polymerase chain reaction using fluorescently labeled primer amplifies seven microsatellite markers—five mononucleotide (NR-21, BAT-26, BAT-25, NR-24, and MONO-27) and two tetranucleotide (PENTA C and PENTA D) repeats.
NGS was performed using the Columbia Comprehensive Cancer Panel. The CCCP targets exonic and intronic sequences in 467 cancer-associated genes using DNA from formalin-fixed, paraffin-embedded tissue using Custom Agilent (Santa Clara, Calif.) SureSelect capture and Illumina (San Diego, Calif.) HiSeq 2500 sequencing. Sequences were aligned and nucleotide variants were called using NextGENe (SoftGenetics, State College, Pa.) software. Nucleotide variants were manually curated and classified after filtering with an in-house pipeline.
- Roh YH, Lee HW, Kim MC, Lee KW, Roh MS. Collision tumor of the rectum: a case report of metastatic gastric adenocarcinoma plus primary rectal adenocarcinoma. World J Gastroenterol. 2006;12(34):5569–5572.
- Naka T, Hatanaka Y, Marukawa K, et al. Comparative genetic analysis of a rare synchronous collision tumor composed of malignant pleural mesothelioma and primary pulmonary adenocarcinoma. Diagn Pathol. 2016;11:38.
- Deng G, Bell I, Crawley S, et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10(1):191–195.
- Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793.
- Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J Mol Diagn. 2017;19(2):187–225.
[hr]
Test yourself
Here are three questions taken from the case report.
Answers are online now at www.amp.org/casereports and will be published next month in CAP TODAY.
1. Which of the following are included in the differential diagnosis of spindle cell lesions of the gastrointestinal tract?
a) Sarcomatoid carcinoma
b) Gastrointestinal stromal tumor
c) Metastatic leiomyosarcoma
d) Sarcomatoid mesothelioma
e) All of the above
2. Which of the following is not a gene that encodes for a mismatch repair protein?
a) MLH1
b) MSH2
c) MSH6;
d) BRCA2
e) PMS2
3. Activating mutation in the BRAF gene in colorectal carcinoma correlates most closely with which of the following?
a) Hypermethylation of the MLH1 gene promoter
b) Somatic loss-of-function mutation in the MLH1 gene
c) Germline loss-of-function mutation in the MLH1 gene
d) Loss of heterozygosity at the MLH1 gene locus
[hr]
Dr. Heymann, Dr. Mansukhani, Dr. Sireci, and Dr. Sepulveda are in the Laboratory of Personalized Genomic Medicine, Department of Pathology and Cell Biology, Columbia University Medical Center (CUMC), New York, NY. Dr. Kiran is in the Department of Surgery, CUMC. Dr. Yang is in the Department of Pathology and Cell Biology, CUMC.