Amy Carpenter Aquino
October 2019—How well can analytics predict the risk for sepsis? T. Scott Isbell, PhD, DABCC, director of clinical chemistry and point-of-care testing, SSM Health, Saint Louis University Hospital, at this year’s AACC annual meeting shared his hospital’s experience with Epic’s sepsis predictive tool. Launched in 2017, the tool uses predictive analytics to produce a sepsis score for admitted patients based on regular scans of key data elements in the electronic health record.
Saint Louis University Hospital previously used a systemic inflammatory response syndrome (SIRS) criteria alert system. “That thing fired all the time. Pretty much everybody presenting to our hospital triggered it,” said Dr. Isbell, who is also associate professor of pathology and pediatrics at Saint Louis University School of Medicine. It was no surprise given that the SIRS criteria are not specific to sepsis, he said. “Everybody got pop-up fatigue.”

Dr. Isbell
Epic’s sepsis predictive tool was based on an examination of 350 data elements in a training data set of more than 400,000 patient encounters at three hospital sites. “The model itself runs in the background. It’s always on in our hospital, and it’s set primarily to look at patients either in the emergency department or on the floor in the inpatient units,” Dr. Isbell said. The hospital has used the tool since early this year.
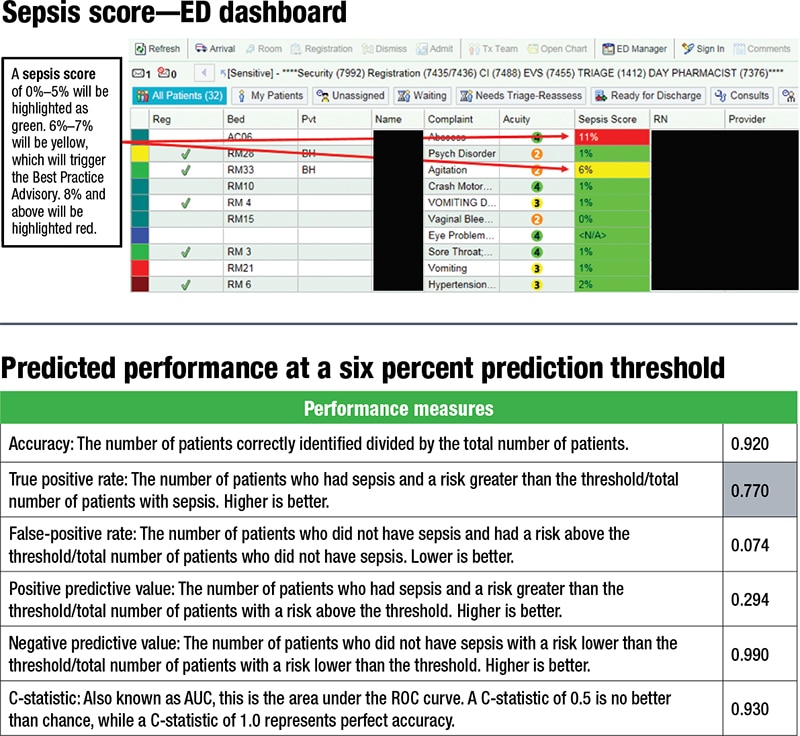
Once a patient is in a room, the Epic sepsis predictive tool begins to scan the patient’s EHR every 15 minutes for key data elements. It feeds the data into the model and generates a sepsis score. Scores of six percent or higher in the emergency department, or eight percent or higher in an inpatient room, trigger a best practice advisory. A pop-up alert indicates that the patient has a risk of developing sepsis—“not a diagnosis of sepsis,” Dr. Isbell said—and requires the clinician to open the order set if the patient’s symptoms are consistent with sepsis. The clinician also has the option to indicate through the alert that the patient’s symptoms are not consistent with sepsis, that further assessment is needed, or that appropriate treatment is already in place. Consulting physicians can defer action to the treating provider.
The Epic tool examines several parameters to generate a sepsis score: demographics; traditional vital signs (which are also the SIRS criteria), such as temperature, heart rate, and respiration rate; comorbidities, such as congestive heart failure, diabetes, and hypertension; and active lines, drains, or airways, such as feeding tubes and peripheral IVs.
The laboratory list of parameters is “fairly short,” Dr. Isbell said. The tool examines mostly hematologic parameters, such as white blood cell count with limited differentiation, particularly lymphocytes, monocytes, neutrophils, and segmented and band neutrophils. It also examines RBC, Hct, Hb, MCHC, nRBCs, RBC morphology, RDW, reticulocytes, and platelet count. Metabolic laboratory measures are creatinine, hemoglobin A1c, base excess, and procalcitonin.
“Glaringly obvious and remiss is that lactate is not on there, which is interesting,” Dr. Isbell said. “It would require digging down, and apparently they didn’t find an association.”
The Epic tool also looks for the presence of various drugs.
The tool’s sepsis score tracker recalculates the patient’s sepsis score every 15 minutes. “It’s dynamic,” he said. “It’s not a one-time score.”
Epic’s performance results showed a positive predictive value of 16 percent with a threshold set at four percent, and a positive predictive value of 50 percent if the prediction threshold is set at 40 percent, according to an Epic report. “The negative predictive value was very similar, irrespective of where you are,” he said: 93 percent with a prediction threshold of four percent and 97 percent with a threshold of 40 percent. “I’m not surprised by this. This is trying to optimize the model in such a way where we are not missing anybody and no one is falling through the cracks.”
But the specificity needs improvement, Dr. Isbell said, noting that it’s unclear at this point if treatment of patients classified as likely to have sepsis is harmful or contraindicated. “It is an active area of research for us at Saint Louis University Hospital and the sepsis committee I sit on.”
A tool can be used to “play around with the threshold,” Dr. Isbell said. If he and colleagues set theirs at six percent, they’re expecting to see a positive predictive value similar to what was seen at 30 percent, and a negative predictive value at 99 percent. “This tool allows for some level of customization depending on desired outcomes and patient populations.” (See “Predicted performance at a six percent prediction threshold.”) “What we’re trying to do now is collect the actual data and see how in real life does this stack up to what our predictive performance would be.”
He and colleagues are monitoring the sepsis alert firing frequency by location and the response rate—the number of alerts responded to within 30 minutes. “We’re probably at 50 to 60 percent, so we have more work to do in terms of people acting on that order set. A teaching hospital complicates that because new people come in and move around.”
They’re also monitoring the SEP-1 outliers. “Are we adhering to the model, are we ordering blood culture, are we giving them fluids? Are we doing all these things like we’re supposed to?
“To summarize, I think predictive analytics is a tool that can help identify patients likely to become septic,” Dr. Isbell said, adding that the tool needs fine-tuning. “Our main goal is to be able to predict with a high degree of sensitivity and specificity those likely to develop sepsis, treat them, and prevent them from developing or advancing to septic shock.”
Amy Carpenter Aquino is CAP TODAY senior editor. Amanda Thompson, RN, BSN, patient safety and quality coordinator and chair of the SSM Health SLU Hospital sepsis committee, provided some of the data and other information Dr. Isbell presented at the AACC meeting. Dr. Isbell presented in the same session as David Ford, PhD; see story.