CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from National University Health System in Singapore. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Gabriel Yan, MBBS; Chun Kiat Lee, MSc; Shaun Tan, MBBS; Stephen Chew, MBBS; Paul Anantharajah Tambyah, MBBS; Benedict Yan, MBBS
December 2021—Clinical metagenomic next-generation sequencing (mNGS), the comprehensive analysis of microbial and host genetic material (DNA and RNA) in patient samples, is increasingly available in clinical laboratories.1 At present, for various reasons including test complexity, cost, and turnaround time, this technology is generally limited to difficult-to-diagnose conditions where conventional microbiological testing methods may not lead to a definitive diagnosis in selected patients, such as those with meningoencephalitis of unknown etiology.2
Encephalomyocarditis virus (EMCV) is a group of closely related virus strains with a wide host range.3 First isolated by Helwig and Schmidt in 1945,4,5 EMCV infection is associated with sporadic cases and outbreaks of myocarditis and encephalitis in many non-human mammals. Disease transmission is poorly understood, and rodents, and more recently bats, are thought to be the natural reservoirs.6 A serological survey in Peru showed the prevalence of human exposure to range from six percent to more than 17 percent.7 In this report, we describe the identification of EMCV from metagenomic analysis of urine from a patient with an acute febrile illness.
Case. A 56-year-old female Singaporean patient who had traveled to Madagascar from Sept. 29 to Oct. 15, 2019 developed fever, nausea, and myalgia five days prior to her departure from Madagascar. There were no other associated symptoms, and physical examination was unremarkable. She had previously been on doxycycline prophylaxis for malaria.
The fever abated on day seven of her illness, but the malaise persisted, and a complete blood count performed was unremarkable. Of note, the platelet counts were within normal reference limits (285 × 109/L). A rapid dengue test performed on that day was positive for dengue IgM and negative for NS1 antigen and dengue IgG (Wells Bio CareUS Dengue Combo, South Korea). Additional urine and blood samples also tested negative for dengue and Zika virus by reverse transcriptase-polymerase chain reaction.8
Due to the travel history, normal blood counts, and negative PCRs, the positive dengue IgM was deemed a likely false-positive result, and an unbiased mNGS assay was performed from the patient’s urine on day eight of illness. (Urine was chosen as the specimen type because one of the differentials was Zika virus infection, which is detectable on urine PCR.)
Total nucleic acids were purified from 400 µL of urine using the Qiagen EZ1 Advanced XL. To identify RNA viruses, randomly amplified cDNA was generated from RNA (in the total nucleic acid extract) via first-strand and second-strand cDNA synthesis, followed by amplification based on a previously described primer-extension pre-amplification methodology.9 To identify DNA viruses, the DNA in the total nucleic acid extract was used for the library preparation. The Illumina Nextera XT DNA Library Prep Kit was used for both cDNA and DNA library preparation according to the manufacturer’s instructions. Next-generation sequencing of the cDNA and DNA library was then performed using the Illumina MiSeq platform.
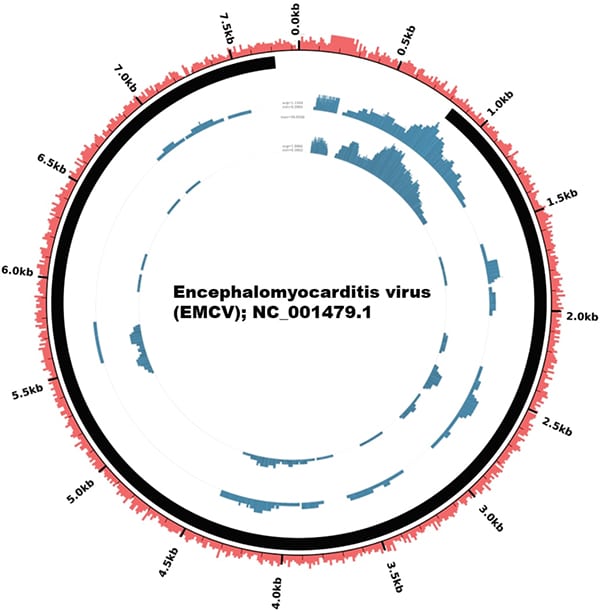
Fig. 1. Genome coverage plot of the EMCV sequenced from the patient’s sample showing 86 percent coverage of the entire EMCV genome. Note that the viral genome (7,835 bp linear RNA) is depicted as circular in this figure as the plot was generated using the Circleator software, an open-source tool that generates circular visualization of genome-associated data. The sample was sequenced in duplicate. Each concentric circle represents a genome coverage plot of the virus. Percentage GC content, read coverage, and viral polyprotein are annotated in red, blue, and black, respectively.
Paired-end reads were analyzed using SUVIA (Sensitive Unbiased Virus Identification Accelerated), a laboratory-developed bioinformatics pipeline that involves quality-trimmed reads mapped against a database of viral nucleotide and protein reference sequences from 10,860 viruses. Of 934,322 reads that were generated from the sequencing, 544 (0.06 percent) mapped to the encephalomyocarditis virus. Of note, there were no reads mapping to dengue virus or other known viruses. Subsequently, reads that mapped to EMCV were retrieved and remapped against the reference EMCV sequence (NC_001479.1) to create the genome coverage plot using the Circleator software, an open-source tool that generates circular visualization of genome-associated data.10 Notably, the reads mapped to different regions within the EMCV genome with 86 percent coverage (Fig. 1), which lends greater confidence to the specificity of the result (i.e. that the result is not a false-positive result). Orthogonal confirmation of the presence of EMCV was performed using specific primers designed to target the 5’ untranslated region of the EMCV genome by RT-PCR,11 which yielded a positive result from the patient’s urine. Phylogenetic analysis revealed that, among 65 EMCV genomes, our patient’s sample was closest to isolates sequenced from swine from Cyprus, Italy, and Belgium (Fig. 2). The percentage sequence difference between our patient’s sample and those from swine from Cyprus, Italy, and Belgium ranged from 18 to 19 percent.
The patient’s symptoms had largely resolved by the time the sequencing results were out, and as such the identification of EMCV as the cause of the acute febrile illness did not have any clinical impact.
Discussion. This report illustrates the potential of mNGS to resolve infections of uncertain etiology where routine methods have been unsuccessful or suspect. In this case, the relatively normal platelet levels on day seven of illness with a negative dengue PCR result were highly suggestive that the dengue IgM result was likely a false-positive one. mNGS of the urine on day eight of illness revealed the presence of EMCV reads, which was confirmed by PCR from the urine; this strongly suggests EMCV as the cause of the patient’s acute febrile illness.
The literature concerning human EMCV infection is sparse. Based on a limited number of recent studies, the common clinical manifestations of human EMCV infection are fever, malaise, nausea, and headache,3,7 consistent with our patient’s clinical presentation. Cardiac complications are apparently rare, having been described in only one patient.7 EMCV infection has previously been described in Singapore, though as fatal cases of myocarditis in orangutans in the Singapore Zoological Gardens, and not in humans.12
With regard to the likely false-positive dengue IgM results, the dengue rapid test uses lateral flow immunochromatography, where anti-dengue IgM and/or IgG to dengue virus in the specimen reacts with antibodies mobilized on the test membrane. False-positive dengue IgM tests due to cross-reactivity with other non-dengue pathogens have been reported.13
mNGS allows for an unbiased diagnostic platform for the detection of pathogens, especially novel or infrequently encountered organisms where traditional methods are unable to identify them, as this case highlights. Typically, where mNGS does lead to the detection of the causative pathogen, only a very small percentage of the total number of reads is attributable to the pathogen, as seen in our case and others.2 This is due in part to the vast majority of reads belonging to the human host genome.1 Importantly, the mNGS results need to be confirmed by an orthogonal clinical-grade assay such as RT-PCR.1
Other known limitations of mNGS include the fact that sequencing only identifies genomes, which does not mean the organism detected is viable. Additionally, the sensitivity of the assay means “noise” will be detected in the form of contaminated genomic material from reagents or from the laboratory environment, which can potentially lead to false-positive results.1 One mitigating strategy is to maintain a database of background microorganisms commonly detected in mNGS data and arising from normal flora or laboratory contamination.1 In our case, such noise was not encountered to any appreciable degree.
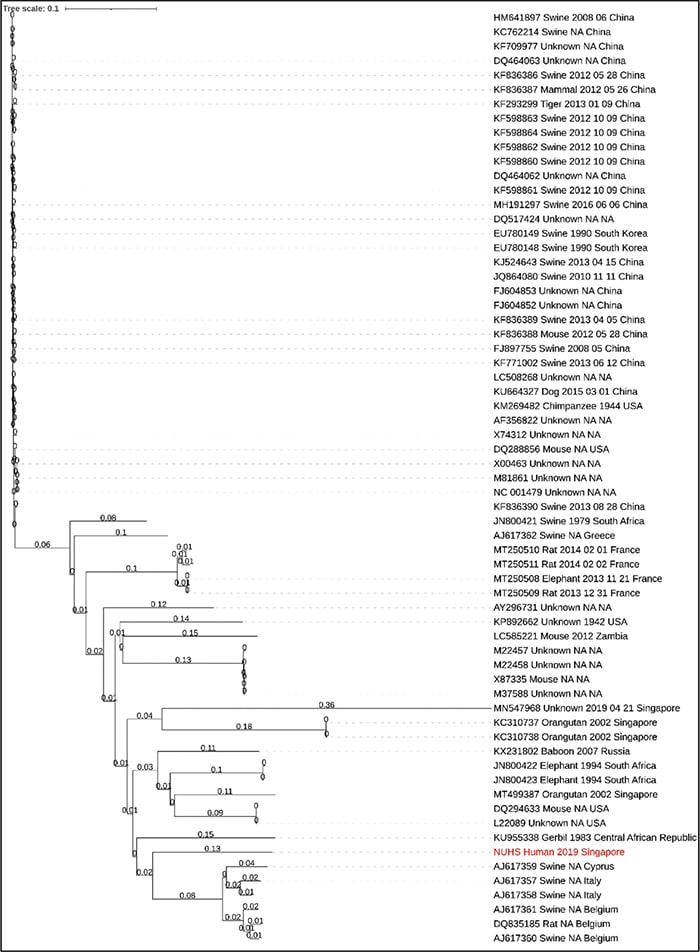
Fig. 2. Bio Neighbor-Joining (BioNJ) phylogenetic analysis of 65 EMCV genomes. Our patient’s sample (red font) was closest to isolates sequenced from swine from Cyprus, Italy, and Belgium.
To our knowledge, this is the first report of EMCV diagnosed through metagenomic analysis. The patient’s travel history, together with atypical blood tests for dengue infection despite a positive dengue IgM serology, led to the conclusion of a false-positive dengue serology result, especially once EMCV was identified in the urine. This case highlights the role of metagenomics in the clinical laboratory for characterization of unknown pathogens, especially in that of a returning traveler.
- Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–355.
- Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–2340.
- Oberste MS, Gotuzzo E, Blair P, et al. Human febrile illness caused by encephalomyocarditis virus infection, Peru. Emerg Infect Dis. 2009;15(4):640–646.
- Helwig FC, Schmidt CH. A filter-passing agent producing interstitial myocarditis in anthropoid apes and small animals. Science. 1945;102(2637):31–33.
- Carocci M, Bakkali-Kassimi L. The encephalomyocarditis virus. Virulence. 2012;3(4):351–367.
- Doysabas KCC, Oba M, Furuta M, et al. Encephalomyocarditis virus is potentially derived from eastern bent-wing bats living in East Asian countries. Virus Res. 2019;259:62–67.
- Czechowicz J, Huaman JL, Forshey BM, et al. Prevalence and risk factors for encephalomyocarditis virus infection in Peru. Vector Borne Zoonotic Dis. 2011;11(4):367–374.
- Lura T, Su T, Brown MQ. Preliminary evaluation of Thermo Fisher TaqMan Triplex q-PCR kit for simultaneous detection of chikungunya, dengue, and Zika viruses in mosquitoes. J Vector Ecol. 2019;44(1):205–209.
- Greninger AL, Naccache SN, Federman S, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi.org/10.1186/s13073-015-0220-9.
- Crabtree J, Agrawal S, Mahurkar A, Myers GS, Rasko DA, White O. Circleator: flexible circular visualization of genome-associated data with BioPerl and SVG. Bioinformatics. 2014;30(21):3125–3127.
- Qin S, Underwood D, Driver L, Kistler C, Diallo I, Kirkland PD. Evaluation of a duplex reverse-transcription real-time PCR assay for the detection of encephalomyocarditis virus. J Vet Diagn Invest. 2018;30(4):554–559.
- Yeo DSY, Lian JE, Fernandez CJ, et al. A highly divergent Encephalomyocarditis virus isolated from nonhuman primates in Singapore. Virol J. 2013;10:248.
- Hunsperger EA, Duarte dos Santos CN, Vu HTQ, Yoksan S, Deubel V. Rapid and accurate interpretation of dengue diagnostics in the context of dengue vaccination implementation: viewpoints and guidelines issued from an experts group consultation. PLoS Negl Trop Dis. 2017;11(9):e0005719.
Dr. Gabriel Yan is associate consultant, Division of Microbiology; Chun Kiat Lee is principal medical technologist, Molecular Diagnosis Centre; Dr. Tan is registrar, Division of Clinical Chemistry; Dr. Chew is senior consultant, Department of Obstetrics and Gynecology; Dr. Tambyah is senior consultant, Division of Infectious Diseases; and Dr. Benedict Yan is a pathologist in the Molecular Diagnosis Centre, Department of Laboratory Medicine—all at the National University Health System, Singapore.
Test yourself
Here are three questions taken from the case report.
1. Which genus among the family of Picornaviridae does the EMCV belong to?
a. Aphthovirus.
b. Avihepatovirus.
c. Cardiovirus.
d. Enterovirus.
2. The common clinical manifestations in human EMCV infection include all of the following except:
a. Fever.
b. Nausea.
c. Malaise.
d. Ascending paralysis.
3. Clinical metagenomics refers to the comprehensive analysis of:
a. Microbial and host genetic material (DNA and RNA) in patient samples.
b. Antibodies in patient samples.
c. Proteins in patient samples.
d. Metabolites in patient samples.
Answers are online now at www.amp.org/casereports and will be published next month in CAP TODAY.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
