CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from the Hospital of the University of Pennsylvania. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Salvatore F. Priore, MD, PhD
Jason N. Rosenbaum, MD
Jacquelyn J. Roth, PhD
 February 2019—Lung cancer is the second most commonly diagnosed malignancy and results in the most cancer-related deaths each year in the United States, but actionable aberrations in EGFR, ALK, ROS1, and other oncogenes are improving outcomes for a subset of patients. The recent clinical practice guideline published by the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology provides guidance in the best molecular testing practices for lung adenocarcinoma specimens: genes that must be tested (EGFR, ALK, ROS1), genes that should be included if an expanded panel is used (BRAF, MET, RET, ERBB2 [HER2], and KRAS), and genes currently considered investigational.1 Activating disease-associated variants in the must-test oncogenes EGFR, ALK, and ROS1 increase cell proliferation and tumor cell survival. Despite their comparatively low prevalence in 10 to 35 percent, one to three percent, and one to two percent of lung adenocarcinomas, respectively, they are clinically important because they predict response to therapy with specific receptor tyrosine kinase inhibitors.1-6
February 2019—Lung cancer is the second most commonly diagnosed malignancy and results in the most cancer-related deaths each year in the United States, but actionable aberrations in EGFR, ALK, ROS1, and other oncogenes are improving outcomes for a subset of patients. The recent clinical practice guideline published by the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology provides guidance in the best molecular testing practices for lung adenocarcinoma specimens: genes that must be tested (EGFR, ALK, ROS1), genes that should be included if an expanded panel is used (BRAF, MET, RET, ERBB2 [HER2], and KRAS), and genes currently considered investigational.1 Activating disease-associated variants in the must-test oncogenes EGFR, ALK, and ROS1 increase cell proliferation and tumor cell survival. Despite their comparatively low prevalence in 10 to 35 percent, one to three percent, and one to two percent of lung adenocarcinomas, respectively, they are clinically important because they predict response to therapy with specific receptor tyrosine kinase inhibitors.1-6
Chromosomal rearrangements, such as ALK and ROS1, can be challenging to detect, owing to the considerable variability of breakpoints and fusion partners. Break-apart ALK and ROS1 fluorescence in situ hybridization addresses this concern by detecting rearrangement at the ALK and ROS1 loci (irrespective of the fusion partner), whereas immunohistochemistry detects ALK or ROS1 proteins (also agnostic of the fusion partner), which can identify aberrant expression in adult lung tissue. Both assays are approved by the Food and Drug Administration as companion diagnostic assays and are recommended as equivalent screening methods for identifying ALK rearrangements in the CAP/IASLC/AMP guideline. In comparison with massively parallel sequencing (MPS)-based panels, IHC and FISH are simple, rapid, and inexpensive; however, they are difficult to multiplex beyond a few targets (particularly on limited or low-quality tissue specimens) and suffer from technical and interpretive limitations. This case highlights several important preanalytic and analytic considerations when testing for rare disease-associated variants in lung adenocarcinomas.
Case. A 68-year-old Caucasian woman presented to her physician in 2017 with new onset cough and shortness of breath. Her past medical history was significant for a Clark level III, Breslow depth 0.5-mm melanoma on her right calf treated by wide excision in 2013. The malignant cells were positive for Melan-A by immunohistochemistry, and the surgical margins were free of tumor. Imaging of the chest revealed a left pleural effusion along with suspicious mediastinal lymphadenopathy, consistent with metastatic disease. Pleural fluid obtained from thoracentesis at an outside institution revealed MART-1–positive and S100- and HMB-45–negative tumor cells. The pathologist diagnosed a malignancy suspicious for melanoma based on the equivocal staining pattern. The patient received ipilimumab and nivolumab therapy for presumptive stage IV melanoma, and testing of archived tissue from 2013 did not detect a BRAF (c.1799T>A, p.V600E) disease-associated variant by pyrosequencing.
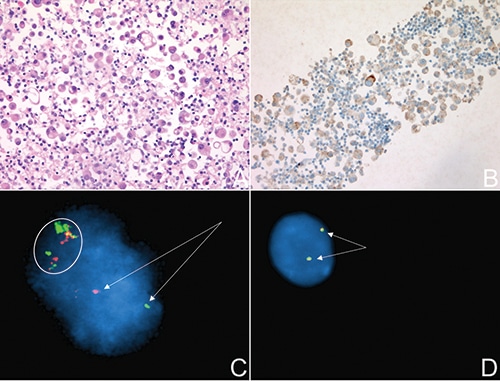
Fig. 1. A. H&E slide of the FFPE tissue produced from the pleural fluid demonstrating a malignant cell population. B. ALK IHC performed on the FFPE tissue shows strong granular cytoplasmic staining in many tumor cells. C. ALK FISH performed on fresh pleural fluid. The arrows indicate the separation of 5′ and 3′ ALK signals consistent with a rearrangement. The separated signals are greater than two probe diameters apart from each other, a validated standard used frequently to identify positive break-apart FISH. The circled area represents colocalized FISH probes with nondiscrete signal separation, suggestive of DNA integrity issues. An additional ALK signal is noted in this one example. D. ALK FISH performed on FFPE tissue from the same pleural fluid. The arrows show colocalization of both ALK signals indicating the absence of a rearrangement. ALK FISH was performed using the Vysis ALK Break Apart FISH probe (Abbott).
The patient sought further management at our institution. Her symptoms did not improve and 15 days later she received a second thoracentesis. IHC performed on the formalin-fixed, paraffin-embedded cytopathology specimen showed tumor cells negative for HMB-45, Melan-A, S100, SOX10, PAX8, GATA3, WT-1, and CDX2. However, the cells were positive for PanCK, CK7, TTF-1, and napsin, supporting the diagnosis of a lung adenocarcinoma rather than metastatic melanoma. The tumor cells tested positive for ALK and negative for ROS1 expression by IHC (Fig. 1A and 1B). Dual-color break-apart FISH performed on an aliquot of the fresh pleural fluid also supported an ALK rearrangement (Fig. 1C). A minimum of six percent of tumor cells must show rearrangement for ALK in pleural fluid specimens, the validated cutoff in our laboratory. Approximately 11 percent of the patient’s cells were positive for ALK rearrangement. The patient began therapy with the ALK-targeted inhibitor alectinib.
Concurrently, a portion of the processed cytology specimen submitted for MPS panels showed EGFR c.2573T>G, p.L858R and TP53 c.818G>A, p.R273H single nucleotide variants (Fig. 2). The turnaround time for MPS results in our lab is about two weeks; therefore, the EGFR result was not yet available when the ALK IHC and ALK FISH results were reported. Although ALK rearrangement and EGFR driver variants have been reported to co-occur in rare cases, they are usually considered to be mutually exclusive. The potentially contradictory results prompted repeat ALK FISH on the FFPE cell block prepared from the pleural fluid sample, as well as testing using a clinical, RNA-based MPS assay designed to directly identify oncogenic fusion transcripts, such as the products of genomic rearrangement involving the ALK locus.7 The panel targets specific exon boundaries of RNA transcripts, enabling multiplexed detection of oncogenic fusions, irrespective of the fusion partner, critical for detecting promiscuous gene rearrangements like those involving ALK. Moreover, because RNA is naturally amplified several orders of magnitude from a single allele, RNA-based sequencing assays can have very high sensitivity. ALK FISH results from the FFPE cell block were wild type (Fig. 1D), and the MPS fusion transcript panel did not detect an ALK fusion transcript. These results prompted a change in therapy from alectinib to EGFR-targeted therapy with osimertinib. Three weeks after initiating osimertinib, the patient’s pleural effusion had largely resolved with excellent symptomatic improvement.
Discussion. As this case illustrates, correctly identifying ALK rearrangements can be difficult. Although ALK rearrangements are often quoted in the range of three to seven percent, these rates are lower in unselected western European population studies (less than one to three percent) but higher in populations with the following characteristics: Asian ethnicity, younger age, never smoking status, advanced clinical stage, and a solid or signet ring cell histology. The patient in this case did present with an advanced clinical stage and was a never smoker, but did not match any of the other associated characteristics.2,8-10
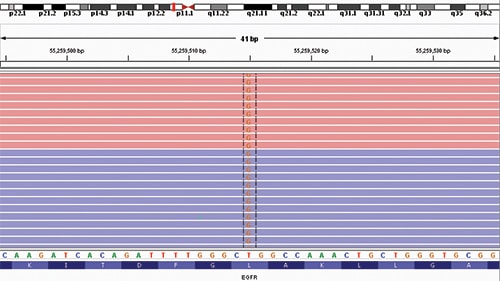
Fig. 2. Integrated Genomic Viewer analysis of EGFR exon 21 showing the c.2573T>G (p.L858R) single nucleotide variant observed in this case. The SNV was detected in eight percent of sequencing reads with a depth of sequencing of 5210 at this position. Red and blue coloring indicate forward and reverse sequencing reads, respectively. A schematic of chromosome 7 is provided at the top of the figure. The reference sequence and predicted amino acid sequence are depicted at the bottom of the figure.
FISH has traditionally been used as the gold standard by which performance metrics for other assays are derived. However, one study comparing ALK rearrangements detected by an MPS panel with ALK FISH on FFPE tissue showed that approximately 18 percent of FISH-positive cases were negative for ALK rearrangements by MPS.11 Another study found that FISH failed to identify several cases in which ALK rearrangement was identified by MPS.12 Some of this discrepancy can be explained by the technical and biological constraints of the FISH assay. The most common ALK rearrangement is caused by an inversion within the short arm of chromosome 2 producing an EML4/ALK fusion. Therefore, the distance between the probes is often still relatively close together within the ALK-rearranged cell. As DNA degrades, this can give the appearance of signals separated in space, when in fact no rearrangement has taken place.13
Depending on the interval between the specimen collection time and FISH testing, a fresh specimen that has been stored unfixed in a refrigerator may be less ideal than an FFPE sample processed soon after receipt. As can be seen in this case, interpretation of the ALK FISH on the pleural fluid clearly shows separation of the probes, but the remaining background signals are quite dispersed, potentially explained by loss of DNA integrity (Fig. 1C). This contrasts with the repeat ALK FISH performed on FFPE from the same specimen (Fig. 1D). Given that FISH probes are dependent on factors such as DNA integrity and secondary structure, it is not unreasonable that normal signals may appear artifactually separated. Proper validation must be performed to differentiate positive versus negative signals under such conditions as well as for the various specimen types used for testing in the clinical laboratory.
In principle, IHC may be more specific than FISH, as ALK protein should not be expressed in normal adult lung; however, even highly specific assays in the setting of a low-prevalence disease state can produce more false-positive results than true-positive results. The sensitivity and specificity of ALK IHC has been reported as 90 percent and 97.8 percent, respectively.14,15 Notably, these data are usually derived by comparison with FISH, which itself is a less than perfect gold standard. Thus, given an ALK rearrangement prevalence rate of one to three percent in all new lung adenocarcinomas, only 31 to 58 percent of cases with a positive IHC result will contain an ALK rearrangement, respectively; the positive predictive value of IHC, and other tests, must always be assessed in the context of the tested patient population. A recent article explores more completely the preanalytical, analytical, and postanalytical variables inherent to immunohistochemistry of pulmonary biomarkers.16
Although no official recommendation currently exists for using either DNA- or RNA-based MPS panels to detect ALK rearrangements, such panels can be a powerful tool, providing orthogonal data to resolve potential false-positive findings when clinical suspicion for an ALK rearrangement is low. One advantage of multiplexed assays is they afford the opportunity to consider known biology and pathogenesis of lung adenocarcinomas. For example, EGFR and ALK have rarely been co-associated and most studies have found them to be mutually exclusive events. Like ALK rearrangements, aberrations in the EGFR gene are enriched in patients with a never smoking status and female gender. Most clinically relevant changes occur in the EGFR kinase domain (exons 18–21). Some changes, such as the c.2573T>G (p.L858R) seen in this case, are known to sensitize lung cancer to targeted TKI therapy (Fig. 2).17 Although tumor heterogeneity is always possible, the patient’s demographics, the much higher prevalence of EGFR variants, the high specificity of DNA-MPS (which identified the EGFR variant), the high sensitivity of RNA-MPS (which did not identify an ALK fusion), and the other ALK testing in aggregate all point to a true EGFR single nucleotide variant by MPS and false-positive ALK rearrangement by FISH and IHC. The patient’s failure to improve on alectinib and excellent clinical response to osimertinib reinforce this conclusion.
Other interesting aspects of this case deserve consideration. About four percent of lung adenocarcinomas will contain activating driver disease-associated variants in the BRAF gene, including p.V600E, and there is a newly designated role for BRAF-targeted therapy in tumors that contain this aberration.18 If it were present, a BRAF p.V600E variant in this tumor might have suggested inappropriately that the disease in the pleural fluid was related to the previous melanoma diagnosis. All metastatic tumors of unknown primary should receive a thorough initial workup. Although this patient did have a history of melanoma, metastasis would be very uncommon given the pathological characteristics of her 2013 resection specimen. Finally, TP53 variants, as observed in this case, are present in about 46 percent of lung adenocarcinomas.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
