CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from the Hospital of the University of Pennsylvania. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Neha Gupta, MD; Selina Luger, MD; Dale M. Frank, MD; Jacquelyn J. Roth, PhD; Salvatore F. Priore, MD, PhD
February 2022—Myeloproliferative neoplasms (MPNs) are a group of clonal hematopoietic stem cell disorders characterized by increased proliferation of myeloid cells of one or more lineage. The most common MPNs include chronic myeloid leukemia (CML), polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). CML is defined by the BCR-ABL1 fusion, which typically results from the t(9;22)(q34;q11.2) rearrangement.1 JAK2, CALR, and MPL alterations are common in non-CML MPNs (the “classical MPNs”), though not exclusive to any one entity. The most common variant in PV is JAK2 p.V617F, a pathogenic gain-of-function variant observed in more than 95 percent of cases. Approximately three percent of JAK2 p.V617F-negative PV cases will show a JAK2 exon 12 variant. PMF and ET show a similar frequency of genetic alterations, with 50 to 60 percent harboring JAK2 p.V617F, 30 percent harboring CALR variants, and five to eight percent of PMF and three percent of ET displaying variants in the MPL gene. The remaining 10 to 15 percent of PMF and ET are considered triple negative and may contain an assortment of rare genomic variants.1,2
A heterogeneous collection of rare myeloid neoplasms with the “ungainly designation of myeloid/lymphoid neoplasms with eosinophilia (MLN-Eo) and rearrangement of PDGFRA, PDGFRB, or FGFR1, or with PCM1-JAK2” (WHO 2017),3 lacks the genetic findings that characterize CML and the other classical MPNs. Instead, these neoplasms harbor characteristic recurrent gene rearrangements involving the abovementioned tyrosine kinase genes. Clinically, constitutive kinase activation drives eosinophilia, which has been a useful organizing principle for this group of rare disorders. However, it has become clear that many patients present without significant eosinophilia and with clinical and pathologic findings that resemble a variety of hematologic neoplasms ranging from myelodysplastic syndromes to CML and the classical MPNs. Therefore, these diseases should be considered as part of the differential diagnosis for MPNs without an identified causative genomic variant.3,4 We present a case that highlights the important roles molecular diagnostics and variant classification play in establishing a correct diagnosis and optimal treatment plan for patients who present with features consistent with an MPN.
Case presentation. A 41-year-old female with a past medical history of surgically managed cervical cancer presented at an outside hospital with cough, dyspnea, congestion, malaise, abdominal pain, sweats, and myalgias. MRI of the abdomen revealed hepatosplenomegaly, liver cysts, and mild retroperitoneal lymphadenopathy. A complete blood count showed a mild leukocytosis, a myeloid left-shift, and nucleated red blood cells. Hemoglobin and platelets were within normal limits.
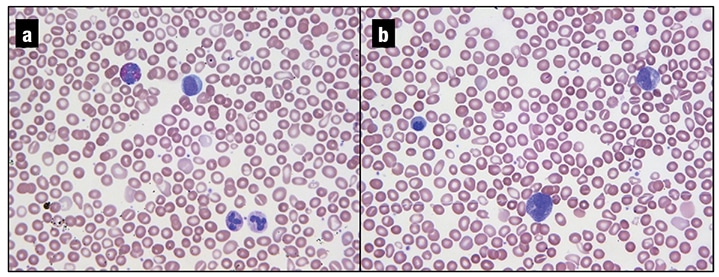
Fig. 1. a and b. Peripheral blood smear, 40×. The leukoerythroblastic picture demonstrated in these images (a mild leukocytosis with a myeloid left-shift, including a few blasts, teardrop cells, and nucleated RBCs) is characteristic of underlying marrow fibrosis.
The patient was referred to a hematologist for further evaluation. A repeat CBC confirmed these abnormalities. Outside bone marrow studies showed morphologic abnormalities (marked hypercellularity, mild megakaryocytic atypia, and grade one reticulin fibrosis) diagnostic of an MPN and favoring primary myelofibrosis. Cytogenetic studies performed on a hemodilute bone marrow aspirate were non-diagnostic; fluorescent in situ hybridization studies were negative for a BCR-ABL1 rearrangement. Molecular studies (real-time PCR followed by melting curve analysis) performed on the bone marrow aspirate were reported as “positive for CALR exon 9 insertion/deletion mutation.” The resulting cDNA and protein changes were reported as “c.1142_1144delAGG” and “p.E381fs,” with an estimated allele fraction (VAF) of 50 percent. All CALR nomenclature in this case report is consistent with reference sequence NM_004343.3. The targeted molecular studies were negative for JAK2 p.V617F, JAK2 exon 12, JAK2 exon 13 (p.G5712S), and MPL W515L/K variants.
The patient was then referred to our institution for a second opinion. Review of a peripheral smear showed a leukoerythroblastic picture with teardrop forms (Fig. 1). CBC findings were similar to those noted previously: WBC 12.9 THO/µL (neutrophils 61.4 percent, bands one percent, metamyelocytes one percent, myelocytes one percent, promyelocytes two percent, lymphocytes 16.8 percent, monocytes 11.9 percent, eosinophils 2.9 percent, basophils one percent, blasts one percent), Hgb 13.3 g/dL, MCV 96 fl, platelets 176 THO/µL. Review of the outside hospital’s bone marrow study confirmed the diagnosis of myelofibrosis and noted mild eosinophilia (eight percent) on the aspirate smear.
Massive parallel sequencing (MPS) studies were performed in our institution’s CAP/CLIA-certified laboratories on a peripheral blood specimen collected one month after the initial outside hospital’s bone marrow biopsy. Our MPS assay is a hybrid-capture–based comprehensive genomic panel designed to detect single nucleotide variants, insertions and deletions, and select copy number gains in 116 genes with therapeutic, diagnostic, and/or prognostic importance for hematological malignancies. Data analysis was performed using a custom bioinformatics pipeline. All variants were annotated with reference to the hg38 genome build. The patient’s sample was positive for one missense variant of uncertain significance in the NFKBIE gene, c.266C>T, p.A89V, with a VAF of 51 percent (NM_004556.2). Sequencing did not detect pathogenic CALR exon 9 deletions, JAK2 exon 12 deletions, JAK2 p.V617F, or MPL codon p.W515 variants.
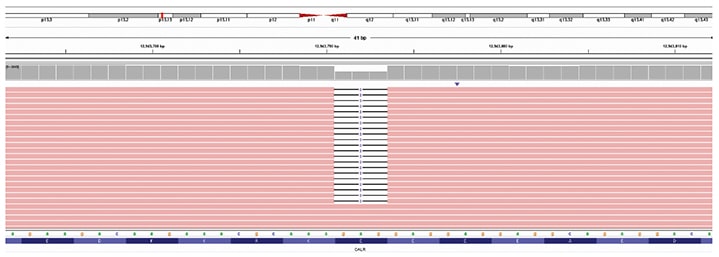
Fig. 2. CALR exon 9 c.1142_1144del (NM_004343.3) variant was detected using massive parallel sequencing. Reference DNA and protein sequences are displayed on the bottom of the figure. The CALR gene position on chromosome 19 (19p13.13) is indicated by the red vertical bar at the top of the figure. Sequencing reads are displayed in the center of the figure as red horizontal bars with deleted nucleotides represented by white spaces. About 50 percent of the sequencing reads contained a 3 bp in-frame deletion.
Given the apparent discrepancy between the initial molecular study, which reported a CALR exon 9 frameshift variant, and the in-house MPS study, which did not, the molecular findings of the outside hospital were reviewed. This revealed an incorrect predicted protein annotation for the reported c.1142_1144AGG variant, which is an in-frame deletion and not a frameshift variant. When we filtered our MPS data for in-frame deletions in CALR, the same c.1142_1144delAGG variant (more concisely described as c.1142_1144del) was identified (Fig. 2). As this variant was internally classified as a benign change, it was not flagged for review or reporting during our variant review workflow.
Given essentially negative MPS results, the referring oncologist ordered additional studies, all of which were performed outside our institution. A rearrangement of JAK2 (9p24.1) was detected in 52 percent of cells (156/300) using an investigational-use-only dual-color breakapart probe set (CytoCell). This result was confirmed by conventional cytogenetic analysis with a 46,XX,t(8;9)(p22;p24)[10]/46,XX[10] karyotype. An investigational-use-only MPS panel (Archer FusionPlex) also detected an in-frame fusion between PCM1 exon 36 (NM_006197) and JAK2 exon 11 (NM_004972).
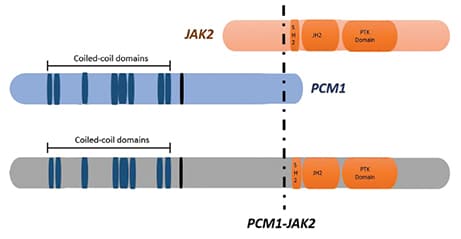
Fig. 3. A schematic representation showing the structure of the PCM1-JAK2 fusion protein.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
