CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from MD Anderson Cancer Center at Cooper, Cooper University Health Care, and Cooper Medical School of Rowan University. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Blair Barnes, MD; Faith Young, MD
Kristin Mattie, MS; Kathryn Zarnawski, MGC
Tina Bocker Edmonston, MD
November 2021—Molecular analysis of advanced stage tumors has become the gold standard for identifying potential targetable mutations with high sensitivity, even in limited size tissue samples. However, when only tumor tissue is sequenced, it is difficult to differentiate between somatic mutations in the tumor cells versus constitutional (germline) mutations.
Case. A 70-year-old Black woman presented with left-sided low chest pain. Imaging revealed a liver lesion. Her past medical history included early-stage breast and lung cancer of unknown histologic subtype (both about 20 years ago), as well as noninvasive low-grade papillary bladder cancer (about two years prior), all amenable to local treatment. At follow-up in the cancer center, the patient also complained of several weeks of intermittent visual disturbances, generalized shaking, bloody stools, and weight loss.
Imaging showed brain metastases, a liver metastasis, and mediastinal and hilar adenopathy. A colonoscopy revealed an ascending colon cancer, which was resected. The brain metastases were treated by radiosurgery, and the liver metastasis was biopsied.
Characterization of the primary colon cancer. The histology of the colon cancer consisted of an adenocarcinoma with tall columnar nuclei forming irregular glands of moderate complexity with focal extracellular mucin, tumor infiltrating lymphocytes, and pushing borders. Immunohistochemistry for the DNA mismatch repair (MMR) proteins MLH1, PMS2, MSH2, and MSH6 was performed according to the NCCN recommendation for universal MMR screening of all colorectal cancers.1 Loss of MLH1 and PMS2 expression in the tumor cells suggested a DNA mismatch repair deficiency (Fig. 1a). MLH1 promoter methylation testing was then performed to determine whether the DNA mismatch repair deficiency was sporadic and caused by somatic gene silencing. The tumor was found to be MLH1 promoter methylation negative, raising suspicion for a possible underlying germline mutation of MLH1 (Lynch syndrome) and thus an inherited predisposition to develop colon, uterine, ovarian, and other cancers. The patient was referred for genetic counseling according to our institutional standard for this scenario, and she decided to pursue germline testing for cancer predisposition (results follow under genetic/germline testing, below).
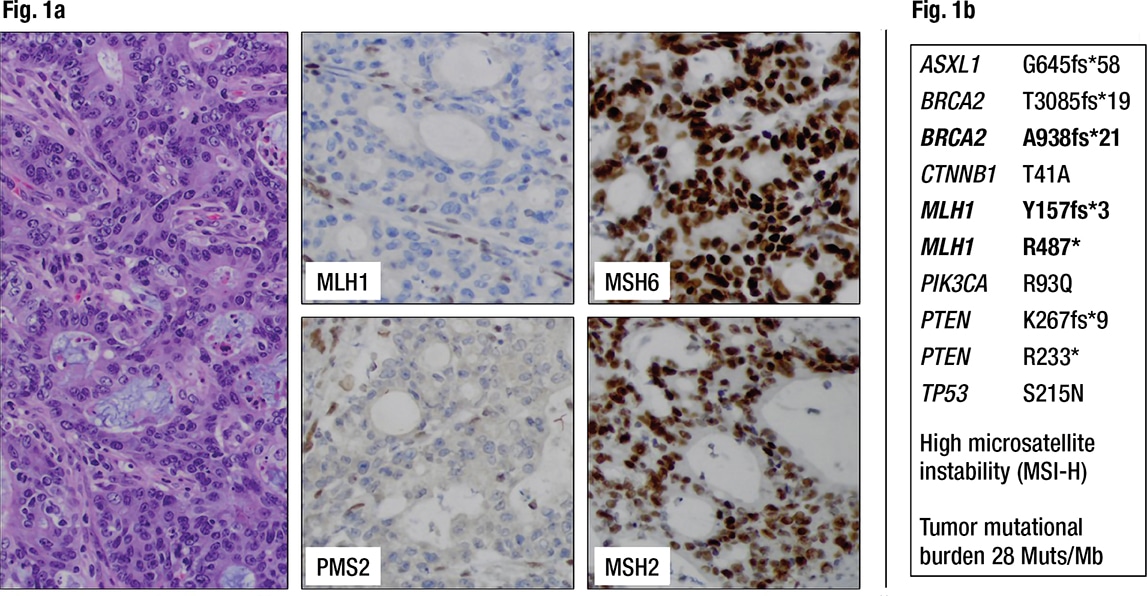
Fig. 1a. Colorectal cancer showing loss of MLH1 and PMS2 expression in the cancer cells but retained nuclear staining for MSH2 and MSH6, indicating a DNA mismatch repair deficiency (dMMR) (200× magnification). Fig. 1b. Next-generation sequencing detects pathogenic variants in the colorectal cancer including two inactivating mutations of MLH1 as well as high microsatellite instability (MSI-H). Frameshift mutations are noted in several other genes, including BRCA2.
In addition, a large next-generation sequencing panel on the colon cancer was requested from a reference laboratory by the oncologist to understand a possible molecular relationship of the colon cancer and the liver lesion. This panel tests only the tumor DNA without comparison to normal tissue, using a hybrid-capture–based library preparation that covers all exons of more than 300 genes as well as selected introns to allow detection of clinically relevant rearrangements. The variant allele frequency (VAF) was not part of the report from this reference laboratory. The colon cancer showed high microsatellite instability (MSI-H) and two truncating MLH1 mutations, p.Y157fs*3 and p.R487*, as well as pathogenic variants in ASXL1, BRCA2, CTNNB1, PIK3CA, PTEN, and TP53 (Fig. 1b). Notably, the majority of variants are frameshifts, possibly caused by the mismatch repair deficiency. Without any additional knowledge about VAF of these MLH1 variants or germline sequence information, the findings of high microsatellite instability and two truncating mutations in MLH1 again raised the possibility of Lynch syndrome.
Characterization of the liver metastasis. Histology of the liver metastasis showed a poorly differentiated adenocarcinoma with pleomorphic nuclei, prominent nucleoli, and areas of gland formation with focal intracellular mucin. It was classified as metastatic high-grade adenocarcinoma consistent with a lung primary based on positivity for CK7, Ber-EP4, and TTF-1, and negativity for CK20, CDX2, villin, GATA-3, PAX-8, uroplakin II, GCDFP-15, and ER (Fig. 2a). However, the immune profile of the patient’s lung cancer in the past was unknown. PD-L1 (clone 22C3) showed high expression (tumor proportion score 95 percent, intensity 2+), which qualified the patient for treatment with pembrolizumab. The lung cancer metastasis in the liver was also analyzed by NGS by the same reference laboratory as the colon cancer, which revealed pathogenic variants in NF1, BRCA2, CDKN2A, and TP53, again without information on the VAF. Of note, this metastasis did not show any MLH1 mutations and was microsatellite stable (Fig. 2b).
Conclusions from molecular characterization of the colon cancer and the liver metastasis from a lung primary. These findings taken together reduced our suspicion of a germline mutation in MLH1. However, the BRCA2 p.A938fs*21 frameshift mutation had been seen in the colon cancer as well as in the lung cancer metastasis, suggesting a possible germline mutation in BRCA2. BRCA2 germline mutations are associated with an inherited predisposition to breast, ovarian, prostate, and other cancers known as hereditary breast and ovarian cancer syndrome (HBOC). However, colorectal cancer and lung cancer are not part of HBOC. Both tumor NGS reports from the reference laboratory mentioned HBOC in a general way in the discussion of the BRCA2 mutation, but did not discuss that in this patient it could be a possible germline mutation based on the VAF or that the same mutation was present in a different tumor of the same patient tested at the same laboratory.
Genetic/germline testing. Germline genetic testing for cancer predisposition mutations in the patient’s blood was initiated based on the MMR deficiency and lack of MLH1 promoter methylation in the colon cancer, before the somatic NGS testing of the liver metastasis had been completed. A reference laboratory tested peripheral blood DNA with an NGS panel of five genes associated with Lynch syndrome and 29 additional genes associated with hereditary cancer predisposition in the germline sample and compared the findings with the MMR gene mutation profile in the colon cancer. This analysis revealed the p.Y157fs*3 and p.R487* variants in MLH1 and copy-neutral loss of heterozygosity of MLH1 only in the colon cancer (somatic) and not in the germline. However, the BRCA2 p.A938fs*21 mutation was identified as a heterozygous pathogenic mutation in the germline sample. This BRCA2 mutation has been reported in multiple families with HBOC and is listed in ClinVar (www.ncbi.nlm.nih.gov/clinvar/variation/9322) where it has been consistently classified as pathogenic by various submitting entities since 1995. Expert panel review on April 22, 2016 supports the classification as pathogenic with the predicted truncated nonfunctional protein (ClinVar, accessed on Aug. 27, 2021). This variant is also listed as a confirmed somatic mutation in two ovarian cancers and one prostate cancer as well as in a lymphoid neoplasm in COSMIC (https://cancer.sanger.ac.uk/cosmic, Genomic Mutation ID COSV66447416, accessed July 9, 2021).
Discussion. The primary goal of somatic biomarker testing of tumor tissue is to detect molecular alterations for the purpose of diagnosis, prognosis, and prediction of treatment efficacy. Different testing modalities are used for different primary tumor types and may include testing for single gene alterations by immunochemistry, fluorescence in situ hybridization, or molecular methods such as RT-PCR or Sanger sequencing. However, while the number of relevant biomarkers has increased rapidly over the course of the past 10 years, the amount of tumor tissue available for testing remains limited.2-4 Therefore, simultaneous testing of multiple genes by NGS, with a relatively high sensitivity (three to five percent VAF for single nucleotide variants and small insertion/deletions in most laboratories using standard protocols for somatic tumor testing), on relatively small tumor samples is gaining popularity and quickly becoming standard of care, especially for lung cancer.5-7
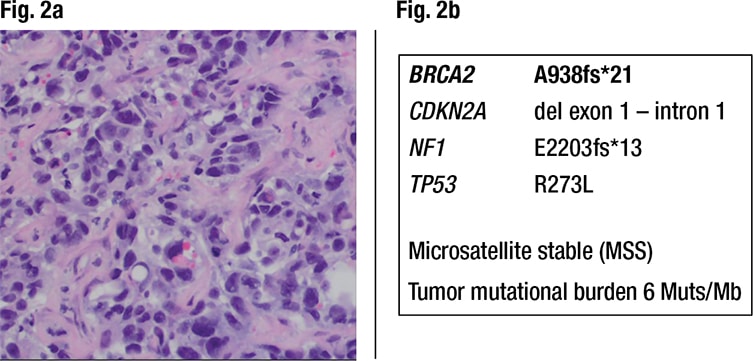
Fig. 2a. Liver metastasis of high-grade adenocarcinoma, consistent with a lung primary (200× magnification). PD-L1 22C3 staining showed a tumor proportion score of 95 percent with 2+ intensity (not shown). Fig. 2b. Next-generation sequencing detects pathogenic variants in the lung cancer metastasis. There is no MLH1 mutation and microsatellites are stable (MSS). The BRCA2 A938fs*21 is present again.
Germline genetic testing is performed on nonmalignant cells of the patient (e.g. peripheral blood, saliva, buccal swabs) with the purpose of identifying a genetic predisposition for certain diseases, including cancer (e.g. Lynch syndrome and HBOC). Germline sequencing of multiple cancer-predisposing genes simultaneously by NGS has become the fastest and most cost-efficient way to test patients given the notable overlap in the clinical presentation of many known cancer predisposition syndromes.1,8,9 One advantage of multi-gene panel testing is that germline mutations can be found in genes that were not expected to be mutated based on the clinical presentation. In a study of 475 patients who were tested with a multi-gene panel, 15.6 percent were found to carry a cancer-predisposing germline mutation but only 47.3 percent of them turned out to be in the genes that the clinicians had previously suspected.10 Possible disadvantages of this testing strategy include an increased frequency of identifying genetic variants of unknown significance or pathogenic variants in genes for which there are no current clinical management guidelines.11,12
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
