CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from Washington University School of Medicine in St. Louis. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Patricia V. Hernandez, MD; Andrea Stacy, MS, CGC
Kevin M. Bowling, PhD; Meagan Corliss, MS, CGC
Yang Cao, PhD
December 2023—The VHL gene, located on chromosome 3, is a tumor suppressor gene that plays a critical role in regulating cell growth and division. Loss-of-function variants in VHL can lead to the development of benign and malignant tumors as well as cysts in various organs, including the central nervous system, kidneys, adrenal glands, and pancreas. The most well-known disease associated with VHL gene alterations is von Hippel-Lindau disease (VHL disease), which is classified as an autosomal dominant disorder although the most common disease mechanism involves the emergence of a second somatic VHL variant in the context of an already altered VHL germline genetic background.1 However, there are few reports in the literature on VHL in the setting of somatic mosaicism.1-3 Here, we present the case of an individual with a clinical diagnosis of VHL disease where next-generation sequence panel testing of peripheral blood and buccal samples yielded a negative germline result, yet a genetic explanation was continually pursued.
Case. A 34-year-old male, with no significant medical and family history, developed spontaneous feelings of dizziness and vertigo associated with occipital pain. The patient underwent further investigation, and computerized tomography of the head showed a 4.2 × 3.8 × 2.8-cm posterior fossa cystic lesion with a mural nodule. Magnetic resonance imaging revealed a 4.7-cm cystic right cerebellar mass with an eccentric enhancing nodule most compatible with hemangioblastoma. Along with this cerebellar lesion, an incidental 1.4-cm × 1.1-cm lesion was found within the interpolar region of the right kidney on CT. Furthermore, in the pancreas, multiple cysts were observed within the mid and distal body and tail ranging from a few millimeters up to 2.5 cm, which most likely represent multiple intraductal papillary mucinous neoplasms according to imaging. A posterior fossa craniotomy resection was performed without complications. Surgical pathology reported a benign neoplasm arranged in a nested to lobulated pattern with high vascularity composed of delicate capillary-sized small vessels; tumor cells presented as round to oval, mostly uniform nuclei with moderate amounts of clear to pale cytoplasm consistent with hemangioblastoma (WHO grade one) with associated reactive changes, including piloid gliosis in the surrounding cerebellar parenchyma. The tumor burden was not informed. Given the constellation of findings (hemangioblastoma associated with cysts in the pancreas and right kidney), the patient was referred to medical genetics with a concern for von Hippel-Lindau disease.
The patient underwent genetic assessment, and a peripheral blood specimen was sent for a Common Hereditary Cancers panel and von Hippel-Lindau syndrome testing in a commercial genetic laboratory. The gene contents of those two panels are listed in Table 1. Genomic DNA obtained from the submitted sample was enriched for target regions using a hybridization-based protocol and sequenced using Illumina technology. All targeted regions were sequenced at a targeted depth of 50×. Reads were aligned to the human reference (GRCh37) sequence, and genetic variants were called and interpreted in the context of a single clinically relevant transcript. A full-gene sequencing and deletion/duplication analysis was performed. Promoters, untranslated regions, and other noncoding regions were not assessed. All clinically significant variants were confirmed by orthogonal technologies. A KIT variant (c.2801A>G, p.His934Arg) of unknown significance was reported, while no pathogenic or likely pathogenic variants in VHL (NM_000551.3) were identified via germline testing.
Given the strong suspicion of VHL disease, von Hippel-Lindau syndrome testing was repeated on a saliva specimen 24 months post-original testing on the same commercial panel using the same VHL gene transcript (NM_000551.3), which was also negative. Subsequently, deep sequencing of the VHL gene was performed in-house using DNA isolated from a formalin-fixed, paraffin-embedded tissue specimen previously collected during cerebellar lesion resection (hemangioblastoma). The hematoxylin-and-eosin–stained section of the tissue block was reviewed to guide the microdissection of areas of viable tumors for DNA isolation. Somatic testing focused on the coding regions of VHL as well as flanking intronic regions (±10 base pairs). Sequencing reads specific to VHL were bioinformatically selected from a target enrichment capture of 177 cancer-related genes. The sequencing of enriched libraries was carried out in multiplex on an Illumina NovaSeq 6000 using a paired-end, 150 bp configuration with an average targeted depth of approximately 2000×. Reads were aligned and variants were called using reference build GRCh37. Variant interpretation and classification were conducted in accordance with established guidelines of the American College of Medical Genetics and Genomics and Association for Molecular Pathology.4 Interestingly, somatic testing revealed a pathogenic VHL variant (NM_000551.3:c.481C>T, p.R161*) in the FFPE specimen at a variant allele fraction of 30 percent (total depth of 1750×) (Fig. 1). Along with negative results of previous germline testing conducted by commercial laboratories, NGS analysis on the FFPE specimen indicates the mosaic origin of this VHL nonsense variant.
Discussion. A nonsense variant in VHL (p.R161*) was identified in the VHL gene at an allelic fraction consistent with a somatic origin in a patient with strong clinical suspicion of VHL disease. VHL is an E3 ligase that functions predominantly as a tumor suppressor.5 The VHL protein forms a ternary complex with transcription elongation factors B and C, which is critical for the stabilization and activity of VHL.6 Genetic variants in VHL that disrupt this complex lead to an unstable VHL protein that is aberrantly degraded.7
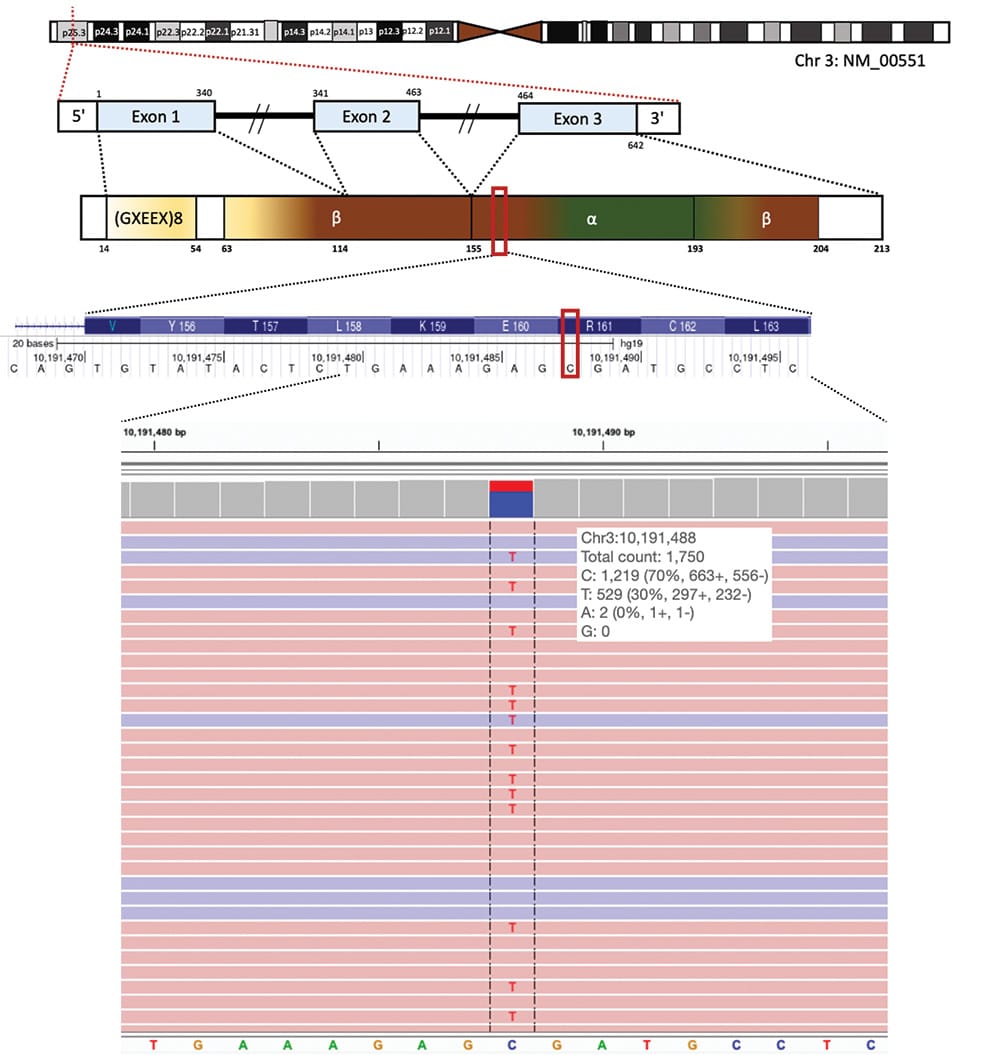
Fig. 1. A somatic VHL variant was identified in exon 3 by NGS testing. The VHL gene locates on chromosome 3 and contains three exons. A nonsense VHL variant (NM_000551.3:c.481C>T, p.R161*) was identified at a variant allele fraction of 30 percent, with the alteration from C to T observed in 529 of 1,750 sequencing reads at chr3:10191488.
The p.R161* variant is predicted to cause loss of normal protein function through protein truncation and subsequently nonsense-mediated mRNA decay, which is an established disease mechanism for VHL-related disorder. The truncated region of the VHL gene encodes the alpha domain, which interacts with the elongin BC complex.5 Experimental studies have shown that missense substitutions in this domain impair protein function in vitro,7 indicating that the amino acid residues deleted by this truncating variant are important for protein function. The p.R161* variant has been submitted as pathogenic by multiple laboratories to ClinVar (ClinVar ID 2217) and is absent from the general population (gnomAD database8).
 Approximately 95 percent of individuals with clinical features of VHL disease harbor an inactivating germline VHL variant, which can be identified by standard molecular testing.9 Acquired somatic pathogenic variants in VHL may give rise to sporadic VHL-type tumors in the absence of other hereditary syndrome-associated tumor characteristics. A study conducted in Denmark described 29 individuals presenting with typical VHL clinical features and fulfilling the international clinical diagnostic criteria based on their diagnostic codes in which no disease-causing VHL variants were detected via germline testing.10 Reasons for disease development in the absence of a germline result may include postzygotic mosaicism, aberrant methylation, genetic abnormalities in other genes resulting in a similar clinical phenotype, or complex or noncoding VHL variants not detectable by the genetic tests employed.1,3 Furthermore, the ELOC gene, also known as TCEB1, is a novel cause of VHL disease and was not evaluated in this patient’s germline analysis.9 Functional studies have shown that ELOC variants can lead to abolished elongin BC complex similar to VHL variants, which ultimately will result in impaired protein function.11 Case reports have described individuals who meet clinical criteria for VHL without a positive VHL germline variant test, but instead presenting ELOC pathogenic variants of germline or somatic origin.9 Somatic mosaicism is a plausible explanation for individuals exhibiting typical VHL disease phenotypes in the absence of a detectable germline variant, although reports of disease-causal somatic VHL variants are quite rare.3,12 Also, a 30 percent allele fraction in the tumor along with negative VHL variants on peripheral blood and saliva may be consistent with mosaicism in the tumor only rather than mosaicism in other tissues, considering the individual presents a typical clinical phenotype of VHL disease (hemangioblastoma associated with kidney and/or pancreas cysts). Since the genetic testing was carried out for clinical purposes, unfortunately we did not test other affected tissues such as kidney and pancreas.
Approximately 95 percent of individuals with clinical features of VHL disease harbor an inactivating germline VHL variant, which can be identified by standard molecular testing.9 Acquired somatic pathogenic variants in VHL may give rise to sporadic VHL-type tumors in the absence of other hereditary syndrome-associated tumor characteristics. A study conducted in Denmark described 29 individuals presenting with typical VHL clinical features and fulfilling the international clinical diagnostic criteria based on their diagnostic codes in which no disease-causing VHL variants were detected via germline testing.10 Reasons for disease development in the absence of a germline result may include postzygotic mosaicism, aberrant methylation, genetic abnormalities in other genes resulting in a similar clinical phenotype, or complex or noncoding VHL variants not detectable by the genetic tests employed.1,3 Furthermore, the ELOC gene, also known as TCEB1, is a novel cause of VHL disease and was not evaluated in this patient’s germline analysis.9 Functional studies have shown that ELOC variants can lead to abolished elongin BC complex similar to VHL variants, which ultimately will result in impaired protein function.11 Case reports have described individuals who meet clinical criteria for VHL without a positive VHL germline variant test, but instead presenting ELOC pathogenic variants of germline or somatic origin.9 Somatic mosaicism is a plausible explanation for individuals exhibiting typical VHL disease phenotypes in the absence of a detectable germline variant, although reports of disease-causal somatic VHL variants are quite rare.3,12 Also, a 30 percent allele fraction in the tumor along with negative VHL variants on peripheral blood and saliva may be consistent with mosaicism in the tumor only rather than mosaicism in other tissues, considering the individual presents a typical clinical phenotype of VHL disease (hemangioblastoma associated with kidney and/or pancreas cysts). Since the genetic testing was carried out for clinical purposes, unfortunately we did not test other affected tissues such as kidney and pancreas.
In the preceding described case study, prior germline genetic testing resulted in negative findings contrary to the patient’s clinical phenotype, which was strongly suggestive of potential VHL disease. Follow-up somatic testing (deep sequencing) of the VHL gene in an affected tissue specimen revealed a pathogenic genetic alteration causal to the patient’s disease. This report of a rare case of potential VHL disease due to somatic mosaicism highlights the value of high-depth NGS testing as well as the importance of conducting genetic testing using disease-relevant tissue sources. Moreover, somatic testing should be considered for patients clinically presenting with VHL disease phenotypes in the absence of detectable germline findings. Combining strong clinical suspicion with genetic testing persistence may prove beneficial (diagnostic) for at least a subset of disease-affected patients.
- Prowse AH, Webster AR, Richards FM, et al. Somatic inactivation of the VHL gene in Von Hippel–Lindau disease tumors. Am J Hum Genet. 1997;60(4):765–771.
- Murgia A, Martella M, Vinanzi C, Polli R, Perilongo G, Opocher G. Somatic mosaicism in von Hippel-Lindau Disease. Hum Mutat. 2000;15(1):114.
- Oldfield LE, Grzybowski J, Grenier S, et al. VHL mosaicism: the added value of multi-tissue analysis. NPJ Genom Med. 2022;7(1):21.
- Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424.
- Schoenfeld A, Davidowitz EJ, Burk RD. A second major native von Hippel–Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc Natl Acad Sci U S A. 1998;95(15):8817–8822.
- Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nat Rev Cancer. 2015;15(1):55–64.
- Couvé S, Ladroue C, Laine E, et al. Genetic evidence of a precisely tuned dysregulation in the hypoxia signaling pathway during oncogenesis. Cancer Res. 2014;74(22):6554–6564.
- Collins RL, Brand H, Karczewski KJ, et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–451.
- Andreou A, Yngvadottir B, Bassaganyas L, et al. Elongin C (ELOC/TCEB1)-associated von Hippel–Lindau disease. Hum Mol Genet. 2022;31(16):2728–2737.
- Binderup MLM, Galanakis M, Budtz-Jørgensen E, Kosteljanetz M, Bisgaard ML. Prevalence, birth incidence, and penetrance of von Hippel–Lindau disease (vHL) in Denmark. Eur J Hum Genet. 2017;25(3):301–307.
- Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860–867.
- Santarpia L, Sarlis NJ, Santarpia M, Sherman SI, Trimarchi F, Benvenga S. Mosaicism in von Hippel-Lindau disease: an event important to recognize. J Cell Mol Med. 2007;11(6):1408–1415.
Dr. Hernandez is a resident in clinical pathology, Dr. Bowling is associate professor of pathology and immunology and technical director of clinical and translational genomics, Meagan Corliss is genetic counselor/genomic coordinator, and Dr. Cao is associate professor of pathology and immunology and director of the laboratory genetics and genomics fellowship—all in the Department of Pathology and Immunology, Washington University School of Medicine in St. Louis. Andrea Stacy is a genetic counselor in the Department of Pediatrics at Washington University School of Medicine in St. Louis.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
