
Jones
And if your conventional identification and your MALDI identification don’t match? “The first thing you should do is go back and make sure the right colony even made it to MALDI,” Jones said. “Sometimes when you’ve got a lot of cooks in the kitchen, this can happen. I’ve done it myself.” If the right colony did make it to MALDI, the next step is resolution of discordant results, which at UNC is done mainly by 16S rRNA gene sequencing.
Over the course of its IVD verifications, the UNC laboratory did more than 731 isolates across over 3,000 spots. Of those, 43 isolates gave no identification, but did give correct identifications over other media and time points. Thirty-three isolate errors were detected, 21 of which were either resolved with current reporting strategies or repeat testing or found not to have fit the morphology to begin with.
“An example here would be if I take a Gram-negative identification card, it gives a result of Citrobacter freundii for a given isolate, and mass spec gives a result of Citrobacter werkmanii. So there’s a couple different ways you might think about this,” Jones said. “You could lump them both into Citrobacter freundii group, report them as such, and move on your way.” The UNC laboratory tends to be fairly conservative about the number of species within a given genus that it reports, “if we feel like we may not be adding clinically relevant information, so in our hands Citrobacter werkmanii would have been reported as Citrobacter species.”
Seven errors did require testing post mass spec; these were with isolates Leuconostoc and Enterococcus casseliflavus and gallinarum. “The bottom line here,” Jones said, “is that in our hands, Leuconostoc would give you the right identification on mass spec, but with these two species of Enterococcus, you might get the right identification, or you might get Leuconostoc as an identification.” They wanted to be able to complete their verification of this group of organisms and ultimately decided they could do that using the mass spec identification and a conventional rapid biochemical tool: PYR. “So, with any of those three mass spec identifications, a PYR-negative result is going to go out as Leuconostoc, and a PYR-positive result will go out as Enterococcus casseliflavus/gallinarum group.”
They had to use this algorithm only seven times in the first year after they went live. “But it does underscore the importance of needing to critically look at the data you collect,” she said. “We had five remaining unexplained errors, but ultimately in our hands, the accuracy of mass spec against all other methods of identification stands at greater than 99 percent.”
Once verification was complete, they still needed to write their laboratory procedures before they could go live. “So we still had some questions to ask,” such as: What would constitute an acceptable result?
With Bruker’s MALDI Biotyper, IVD results are valid down to species level with score results above 2.0. With the Vitek MS, results are presented as percent probabilities. “At 60 percent probability, you’re going to have good identifications,” she said. “A lot of low-discrimination results are going to be things that even 16S is going to have a hard time separating, so they’re going to be things like group C/G strep or Achromobacter xylosoxidans/denitrificans, and even getting to this point is a perfectly good and acceptable report.”
Most percent probabilities in the UNC laboratory are 95 percent or better. “However, at UNC, we’re a very conservative laboratory, and we’ve chosen 80 percent as a percent probability cutoff for acceptable results in our hands.” The one exception: Burkholderia. Because UNC is a cystic fibrosis referral center, it has a large CF population, and many CF patients have Burkholderia. “We know the Vitek MS database is not yet complete for this group of organisms, and while they’re working to complete that, we just evaluate these data very carefully.”
Jones addressed a commonly noted issue regarding MALDI-TOF—namely, that it cannot separate Escherichia coli and Shigella. While there are many possible approaches, she and her team have elected to do the following: “If you have a mass spec identification of E. coli, and the isolate is a lactose fermenter, we report E. coli. If the isolate is a lactose non-fermenter and it’s beta-hemolytic, we still report E. coli. If the isolate is a non-lactose fermenter and not beta-hemolytic, we would then proceed to isolate confirmation, and in our hands this is a Vitek 2, Gram-negative ID card.” Her caveat: “We do not do stool cultures, so if you do, you might want to consider what your other options would be, including some kind of Shigella antisera for a more rapid turnaround.”
Where Streptococcus pneumoniae is concerned, this too is a situation where a lab needs to think about what system it has chosen, Jones said. “If you’re using the Bruker system, the system cannot distinguish between Streptococcus pneumoniae and Streptococcus mitis/oralis, so simply having something like a P disk is a perfectly fine solution here.” The Vitek MS can distinguish these organisms, but because they are so closely related,“there will be the rare time that you will get a slash line or a split identification between Strep pneumoniae and Streptococcus mitis/oralis, and even then we will choose to use a P disk” rather than refire and respot.
The Food and Drug Administration in August 2013 cleared the Vitek MS for reporting of 193 Gram-negative, Gram-positive organisms and yeast. “In your reporting section, you’ll see a big U behind organisms that are not cleared for reporting,” she said. “It’s still perfectly fine for you to go ahead and report these organisms as long as you’ve done your proper verification studies. The only caveat is: For those of you who are already interfaced with your LIS, these organisms won’t cross.” Just a few months after the Vitek MS was cleared, the Bruker device was similarly cleared for reporting of many Gram-negative rods.
As Jones found when implementing MALDI-TOF, “any time you make big changes in your laboratory, you’re going to have stress” among staff members. Her recommendation is to help them understand that their basic microbiology skills are going to be crucial to the success of MALDI, but ultimately “the train’s leaving the station, and you want everybody to be onboard.” Be prepared to hear staff claim, “I can do a rapid test faster than you can do a MALDI identification.”
“They’re right about that for some things,” she said. “So think about things like spot indole or Staphylococcus latex. You’ll need to really think through in your lab what’s going to be right for you.” In the UNC laboratory, they use “all MALDI all the time,” but some other labs use MALDI in conjunction with rapid testing. In her view, “That’s a perfectly fine avenue to go down as well.”
Jones took this opportunity to discuss claims of 30-second MALDI identification. “I kind of came into it thinking I was just going to smear it on there and have my answer a second later,” she said. Rather, she found that using the Vitek MS disposable slide, which is separated into three sections with each section reported as a unit, resulted in a 15-minute identification, which she called “a heck of a lot better than where we’ve come from.”
As far as training went, Jones found that some technologists took to spotting better than others. “We would sit with them and say, ‘Don’t hold your hands this way. Hold your hands this way,’ and kind of give them a visual in words by saying things like ‘icing a cake’ to help them get an idea of what it is we wanted them to do.” At the suggestion of another technologist, they also had a special bench made that holds the slide in a position such that the light shines on it directly from above. “Half the lab uses it. The other half doesn’t. Everyone’s happy,” she said, smiling.
Challenges at the Prep Station arose as well. “Prep is basically where you put in your patient demographics: Mary’s urine is in spot one; Joe’s urine is in spot two,” Jones said. “And you can imagine that if you’re sitting there with a stack of plates, and you’re trying to color in between the lines of the spots, and somebody back here is asking you a question, and someone else is asking you a question over there, and the telephone’s ringing over there, it’s very easy to get off kilter.” The solution: establishing a no-chat zone. “You don’t talk to the spotter, and the spotter doesn’t talk to you, so they stay on task.”
To help cut down on clerical and transcription errors, Jones and her team did what she calls “a bit of organizing on the front end.”
“In our laboratory, we’re organized by benches—blood, urines, and the like—so when we spot a wound, we type a W prior to barcoding the accession ID,” she explained. “When we spot a urine, we type a U. Ultimately, results are collated alphabetically, so all our bloods are together, all our urines are together, all our wounds are together. For those of you who are already interfaced with your LIS, this won’t be an issue, and it’ll go away for us when we’re interfaced, but for right now, it helps.”
Laboratories that implement MALDI should think hard about exactly who will be trained to use it. Because the UNC laboratory reads plates during the day only, it uses MALDI during the day only. In her view, laboratories that read plates on two or three shifts would be well advised to think: “Okay, what am I going to do if I get a questionable result? Am I going to page the director? Am I going to hold the report for the next day? How am I going to handle that?”
What they do at UNC is to flip through their plates when they arrive in the morning without going through the computer, with a goal of trying to get everything on MALDI as quickly as they can. “For the remainder of the day, pretty much, the other techs spot on their own,” she said. “We had to think about: How are we going to keep this organized? How are we going to know where to find these plates as the day progresses and I’ve moved on to do susceptibility testing or something?”
She and her team decided to create cans labeled “mass spec urines,” “mass spec bloods,” and so forth at each bench. After plates are spotted, they go into the appropriate cans and are logged as “culture in progress,” so that if a clinician calls and says, “I’m interested in Mary’s urine,” the technologist knows where to find the relevant plate. And when the MALDI reports come off, they’re entered, but technologists must still match the MALDI result to the plate and proper morphology. “Because even though we establish that no-chat zone back there, well, we all know things happen, right?” Jones said.
Another particularly large issue the laboratory faced in implementing MALDI-TOF was, in Jones’ words, “What things are we now going to put a name to—on a species level, particularly—that we didn’t give a name to before?” Because the UNC laboratory prefers to take a less-is-more approach to avoid overwhelming or confusing clinicians, “Whether or not we were going to add species level to diphtheroids and coagulase-negative Staphylococcus was a big issue for us.”
Ultimately, UNC decided to report the species level only at bloods and sterile fluids for coagulase-negative Staphylococcus. Others, with the exceptions of Staphylococcus lugdunensis and Staphylococcus saprophyticus, are reported simply as coagulase-negative staphylococci.
“We simply sight-read if it’s a skin or respiratory site,” Jones said. “If we didn’t do a tube coagulase or a Staph latex before, we’re not going to do a MALDI now.” Reports would go out simply as probable coagulase-negative Staphylococcus, and they even tack on comments like “skin flora component” where appropriate. “For diphtheroids, we validated and report a limited set, things like Corynebacterium kroppenstedtii, associated with chronic mastitis; Corynebacterium urealyticum, associated with cystitis and urinary calculi; Corynebacterium jeikeium, an opportunistic pathogen that in our hands may or may not be multidrug resistant; and Corynebacterium macginleyi, which is associated with conjunctivitis and corneal ulcers.” Think about citing a reference for some of these new organisms to help clinicians better understand your reports, she advised.
Jones is frequently asked how often UNC’s MALDI system goes down and what happens when it does. “To be quite honest with you, we’re not down a whole lot,” she said. “And the biggest thing that has saved us—and the best advice I can give you—is to simply get comfortable with rebooting the system. In my honest opinion, that takes care of probably 80 percent of any issues we have.”
“Even when you’re down, though,” she added, “you can still use Prep Station to spot your slides the same day and run them up to 48 hours later.”
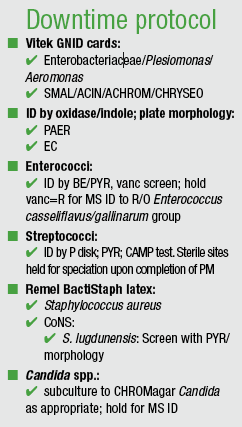
Because annual preventive maintenance for MALDI requires a day to a day and a half of downtime, laboratories that implement it must have downtime procedures in place. “Here’s a snapshot of ours,” Jones said (see “Downtime protocol,” page 6). “This is not comprehensive, but it’s an example. We’ve still got Gram-negative identification cards on hand. We’ve got spot tests on hand. Our technologists know how we’re going to identify enterococci and streptococci. And there are organisms that we’re still going to report as ‘Presumptive ID.’ Sometimes we’re just going to do Gram stain and catalase. Sometimes we’re going to subculture and hold for MALDI, so don’t go live without this in hand, because sure as you do, you’re going to be down the second day—and you’re going to be caught unaware.”
[hr]
Anne Ford is a writer in Evanston, Ill.
![]()
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
