Amy Carpenter Aquino
October 2020—B-ALL with aberrant expression of myeloid markers should be investigated further for specific gene abnormalities, including ZNF384 rearrangements, and microarray analysis may play an important role.
That was the crux of a case presented at last year’s AMP annual meeting by Shweta Bhavsar, MBBS, MD, molecular genetic pathology fellow, University of Pittsburgh Medical Center and School of Medicine.
A second presentation in the same session, by Jeffrey SoRelle, MD, of the University of Texas Southwestern, was of an aggressive case of MDS in which molecular analysis alone led to earlier treatment for the patient.

Dr. Bhavsar
Drs. Bhavsar and SoRelle spoke with CAP TODAY recently.
In the case of B-lymphoblastic leukemia, the four-year-old female patient, who had a significant medical history of asthma, presented with familiar B-ALL symptoms: pallor, fatigue, weight loss, and petechiae, predominantly on her face, Dr. Bhavsar says. “She was not doing well.”
The CBC and peripheral blood exam results revealed leukocytosis (26.3 × 109/L) with approximately 74 percent circulating blasts, and anemia (Hb 8.2 g/dL). “Morphologically the blasts were typical lymphoblasts with no specific findings,” Dr. Bhavsar says, describing them as small to intermediate in size, with a high nucleus-to-cytoplasmic ratio, scant amount of cytoplasm, fine chromatin, and prominent nucleoli.
The bone marrow aspirate results, which showed 92 percent blasts and reduced trilineage hematopoiesis, further supported the diagnosis of B-ALL, she says. “The bone marrow biopsy, unfortunately, was inadequate in this case because it showed predominantly cartilage.”
Flow cytometry revealed that while the blasts, at 86 percent of total events, were positive for CD45 (dim), CD34, HLA-DR, TdT, and the B-cell markers CD19 and CD22, they were negative for CD10, CD20, CD117, cytoplasmic CD3, and myeloperoxidase. “Usually most of the B-ALLs are CD10 positive, but in this case they were CD10 negative, and they showed some myeloid marker expression—CD13 (partial dim) and CD33,” Dr. Bhavsar says. These immunophenotypic findings have been described in association with ZNF384 translocations and some other genetic types, she adds.
On routine karyotypic analysis, five cells showed an inv2 (q11.2q31), which was of unknown significance, as “there is no known specific association of inversion 2 in B-ALLs,” Dr. Bhavsar says. No abnormality was seen on chromosome 12 (ZNF384 is located on Chr 12).
FISH studies—performed for the most common oncogenic alterations in the B-ALL including trisomies 4 and 10, AFF3, BCR/ABL1, KMT2A, ETV6/RUNX1, and CRLF2 gene rearrangements—were all negative.
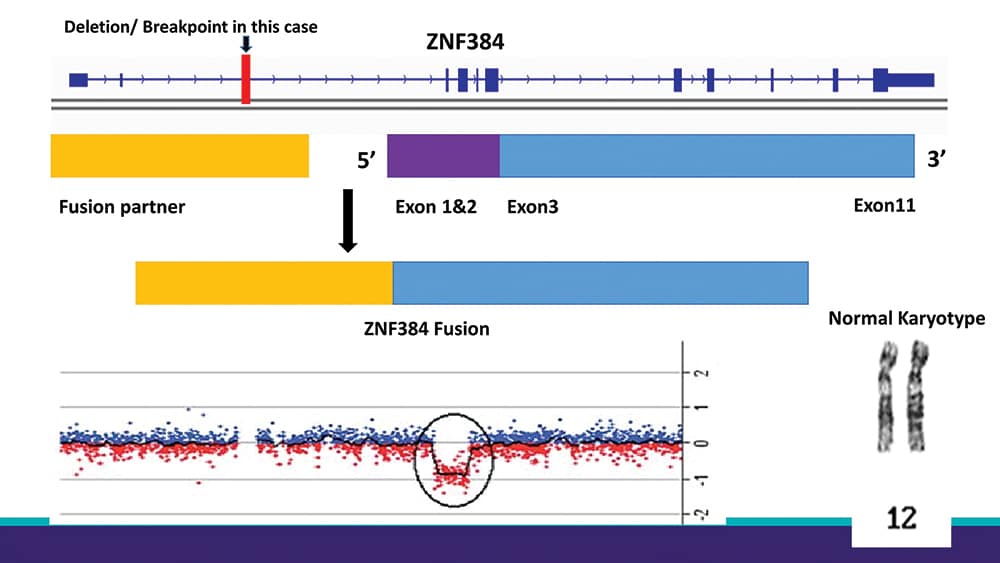
The turning point in the case, Dr. Bhavsar says, was the result of the microarray analysis performed and interpreted by Svetlana Yatsenko, MD, associate professor, Department of Pathology, and director of the Pittsburgh cytogenetics laboratory. (See “Deletion/breakpoint in this case.”)“While balanced translocations cannot be picked up on microarray studies, even when the translocation appears to be balanced, there may be loss of some genetic material at the breakpoints,” Dr. Bhavsar explains. “It is important to review these small deletions in clinically significant genes and look for translocations involving those genes.” This case is an example of such a phenomenon, she says. The microarray results demonstrated a small (62 kb) deletion in the 5′ region of the ZNF384 gene, which is strongly indicative of a translocation involving this gene. Nidhi Aggarwal, MD, hematopathologist and director of UPMC’s molecular hemato-oncology service, Division of Molecular and Genomic Pathology, signed out the morphology on the case. When she reviewed the results of ancillary studies for final classification, she recognized this and further ordered the confirmatory FISH testing. She found recent descriptions of the ZNF384 rearrangement in the literature (Lilljebjorn H, et al. Blood. 2017;130[12]:1395–1401).
“In addition,” Dr. Bhavsar says, “there was a secondary alteration seen in about 20 percent of the cells in chromosome 10, which showed a deletion of the BLNK gene (2.356 Mb), but this was separate from the ZNF384 rearrangement.”
“A lot of people were looking into these B-ALLs, which were initially not categorized in the WHO risk stratification categories,” Dr. Bhavsar says, noting that 20 to 30 percent of B-ALLs do not fall into the established genetic subtypes. “People have been exploring those to see other genetic alterations that could be disease-defining and help to prognosticate and treat patients.”
ZNF384 fusions are one of those new oncogenic subtypes. They have a distinct gene expression pattern that is enriched in hematopoietic stem cell features, “which would explain the aberrant expression of myeloid markers,” she says. ZNF384 fusions in B-ALL have an incidence of about one to six percent in pediatric cases and five to 15 percent in adult cases.
Microarray analysis is a core protocol at UPMC for pediatric B-ALL cases. “Obviously, the main use for microarray is to look for copy number alteration and loss of heterozygosity to recognize hypodiploidy presenting as pseudohyperdiploidy in B-ALL, which can help in prognosis,” Dr. Bhavsar says. “If it’s hyperdiploidy, then these patients do really well, versus hypodiploidy—those patients do badly. So that is one of the key things we look for.”But it can also be a clue to look for these rearrangements if one identifies the small deletions in relevant genes, as in this case, she says. “Otherwise you would have to do specific fusion analysis or RNA sequencing.”
Dr. Aggarwal agrees that microarray analysis is a good screen for copy number and copy neutral changes in any of the genes. For balanced rearrangements, though, it is not a good screen, she says. “As long as the genetic material is present where it is present—whether the rearrangement is present or not—it will not show up on microarray.”
Studies have shown that whenever there are rearrangements—“even rearrangements that we truly think are balanced—at the breakpoint, they actually lose some material. So we think they are balanced but they are not. And that is how we have picked up a few rearrangements in our institute, as demonstrated in this case.”
“That is the important part: Whenever you find these small alterations in microarray, usually the chromosomal analysis does not show anything specific.” If she had not found the ZNF384 rearrangement, Dr. Aggarwal says she would have sent the specimen out for additional testing.
“I was lucky that when I looked at it there was ZNF384 rearrangement, and it was very well described in the literature by that time, although still very new,” she says.
These ZNF384 rearranged acute leukemias are actually a spectrum, Dr. Aggarwal says. While they are described with the B-lymphoblastic leukemias, “they are also described with B/myeloid mixed phenotype acute leukemias or mixed lineage acute leukemias,” she says. “Some people believe that maybe these should just be called as acute leukemia with ZNF384 rather than trying to classify it.”
ZNF384, which has also been known as CIZ and NMP4, encodes a zinc finger transcription factor that is involved in the regulation of matrix metalloproteinases, Dr. Bhavsar says. The exact mechanism of action of ZNF384 fusion proteins is still unknown (Alexander TB, et al. Nature. 2018;562[7727]:373–379).
“One of the unique things about the translocation is the partner gene sequence attaches at the 5′ end of almost the entire gene,” she says. “The entire coding sequence of ZNF384 is present in ZNF384 fusions.”
So far, nine fusion partners have been identified for ZNF384: ARID1B, BMP2K, CREBBP, EP300, EWSR1, SMARCA2, SYNRG, TAF15, and TCF3. “The most common one is TCF3, and there has been a study that talks about how these patients tend to have a poorer prognosis,” Dr. Bhavsar says. “They have a poorer response to steroids and tend to present with higher white blood counts” (Lilljebjorn H, et al. Blood. 2017;130[12]:1395–1401).
Associations with epigenetic regulator genes, such as CREBBP or EP300, have been believed to play a cooperative role in ZNF384 fusions, Dr. Bhavsar says. In in vitro studies, ZNF384 fusions with EP300 and CREBBP have been reported to show increased sensitivity of leukemia cells to histone deacetylase inhibitors. Mutations in the RAS signaling pathway genes (NRAS, KRAS, PIK3CD, PTPN11) are also common (Qian M, et al. Genome Res. 2017;27[2]:185–195).
“So we were able to confirm the ZNF384 rearrangement by FISH and identify its translocation partner to be EP300,” Dr. Bhavsar says of Dr. Yatsenko’s work in the Pittsburgh cytogenetics laboratory. Depending on the partner, “the prognosis of these ZNF384 rearranged cases might vary.”
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
