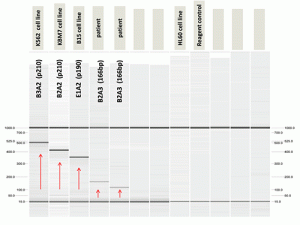
Fig. C. Nested PCR studies for the BCR-ABL1 transcript showed an alternative 166-bp BCR-ABL1 transcript.
CML is a myeloproliferative neoplasm that is associated with the BCR-ABL1 fusion gene in an abnormal myeloid bone marrow stem cell. Approximately 95 percent of CML cases at diagnosis have the characteristic t(9;22)(q34;q11.2) that results in the Ph chromosome. This translocation juxtaposes BCR on chromosome 22 with regions of ABL1 on chromosome 9. The remaining five percent of CML cases show either variant translocations involving chromosomes 9 and 22 with other additional chromosomes or cryptic translocation of 9q34 and 22q11.2. In all cases, the translocation results in the BCR-ABL1 fusion transcript. The transcript has enhanced tyrosine kinase activity and is responsible for constitutive activation of several signal transduction pathways and the leukemic phenotype of CML cells. Elucidation of abnormal signaling in CML cells led to the design and synthesis of small molecule therapies, such as imatinib, that target the abnormal tyrosine kinase activity of the BCR-ABL1 transcript.
Cytogenetic and molecular testing in CML is integral to diagnosis and minimal residual disease (MRD) monitoring. Conventional chromosome analysis is essential for diagnosis and treatment since it establishes the presence of the Ph chromosome in greater than 95 percent of cases. It is also necessary for evaluation of disease progression, as it is the only method that can identify the presence of clonal evolution, which is one of the WHO criteria for CML, accelerated phase. Additional FISH and/or RT-PCR techniques are necessary to identify variant chromosomal abnormalities and establish a baseline BCR-ABL1 fusion transcript level for future MRD monitoring.
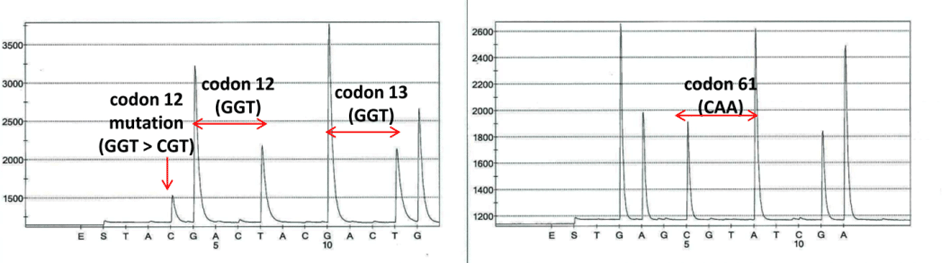
Fig. D. Pyrosequencing analysis of codons 12, 13, and 61 of NRAS. A mutation detected in codon 12 of NRAS (NM_002524: c.34G>Cp.G12R). No mutations detected in codons 13 or 61.
MRD assessment is critical in treatment stratification and prognostication in CML patients. Unlike normal hematopoietic cells, all CML cells harbor the Ph chromosome. Thus, the Ph chromosome is a unique genetic fingerprint that, once detected, forms the basis of MRD monitoring in CML. Conventional cytogenetics can detect the Ph chromosome and other chromosomal changes associated with advanced-phase disease. However, more sensitive molecular techniques such as FISH are needed for MRD testing. FISH is a more sensitive method to detect the BCR-ABL1 fusion and has the advantage of routinely interrogating 50 to 200 metaphase or interphase cells. The most sensitive MRD assay is RT-PCR for the chimeric BCR-ABL1 mRNA. This assay can detect one CML cell in approximately 100,000 to 1 million cells and is the backbone for MRD assessment and clinical management.
Despite the well-documented success of imatinib in chronic-phase CML, imatinib resistance inevitably occurs. This resistance typically results from point mutations in ABL1, which blunt imatinib’s ability to inhibit the aberrant BCR-ABL1 kinase activity. Thus, CML patients receiving imatinib who show either loss of response or disease progression are assessed for ABL1 tyrosine kinase domain mutations via DNA sequencing techniques. If a mutation is present, imatinib is usually replaced with a second- or third-generation tyrosine kinase inhibitor.
Conclusion
Given the frequent morphologic overlap between distinct myeloid neoplasms, a comprehensive clinicopathologic evaluation is essential for precise classification, diagnosis, and treatment. This case emphasizes the significance and utility of multimodal molecular genetic testing to achieve an appropriate diagnosis, prognosis, and therapeutic course in a patient with CML and CMML.
- Borowitz MJ, Bene MC, Harris NL, Porwit A, Matutes E. Chronic myelogenous leukaemia, BCR-ABL1 positive. In: Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer; 2008:32–37.
- Borowitz MJ, Bene MC, Harris NL, Porwit A, Matutes E. Chronic myelomonocytic leukaemia. In: Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer; 2008:76–79.
- Muramatsu H, Makishima H, Maciejewski JP. Chronic myelomonocytic leukemia and atypical chronic myeloid leukemia: novel pathogenetic lesions. Semin Oncol. 2012;39(1):67–73.
- Bacher U, Haferlach T, Schnittger S, Kreipe H, Kröger N. Recent advances in diagnosis, molecular pathology and therapy of chronic myelomonocytic leukaemia. Br J Haematol. 2011;153(2):149–167.
- Baccarani M, Saglio G, Goldman J, et al. Evolving concepts in the management of chronic myeloid leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2006;108(6):1809–1820.
- O’Brien S, Berman E, Bhalla K, et al. Chronic myelogenous leukemia. J Natl Compr Canc Netw. 2007;5(5):474–496.
- Kantarjian H, Schiffer C, Jones D, Cortes J. Monitoring the response and course of chronic myeloid leukemia in the modern era of BCR-ABL tyrosine kinase inhibitors: practical advice on the use and interpretation of monitoring methods. Blood. 2008;111(4):1774–1780.
- Khorashad JS, de Lavallade H, Apperley JF, et al. Finding of kinase domain mutations in patients with chronic phase chronic myeloid leukemia responding to imatinib may identify those at high risk of disease progression. J Clin Oncol. 2008;26(29):4806–4813.
- Fava C, Kantarjian H, Cortes J. Molecular resistance: an early indicator for treatment change? Clin Lymphoma Myeloma Leuk. 2012;12(2):79–87.
Dr. Verma was a molecular genetic pathology fellow, Dr. Wang is an associate professor, Dr. Abruzzo was a professor, and Dr. Sargent was an assistant professor—all in the Department of Hematopathology at UT-MD Anderson Cancer Center, Houston, where Dr. Ferrajoli is an associate professor, Department of Leukemia, Division of Cancer Medicine. Dr. Verma is now a molecular genetic pathologist/hematopathologist at Clarient in Aliso Viejo, Calif. Dr. Abruzzo is now a professor in the Department of Pathology and Laboratory Medicine, Ohio State University Wexner Medical Center, Columbus. Dr. Sargent is now an assistant professor in the Department of Pathology and Laboratory Medicine, The Hospital of the University of Pennsylvania, Philadelphia.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
