- Run 10–20 samples of DNA (or cDNA) from normal FFPE tissue. This establishes false-positives due to sequencing errors, pseudogene interference, or other issues.
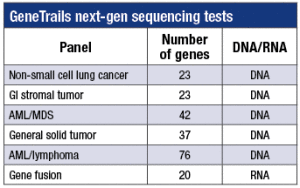
Custom amplicon-based panels validated for DNA (or RNA) from FFPE tissue.
- Run 40–50 samples with known SNVs, including insertions/deletions and/or copy number alterations. The laboratory either uses control cell line samples or FFPE tumor samples with known mutations based on Sanger, MassArray, or other assay. Increasingly, the laboratory is cross-validating from one panel to another using tumor samples with mutations at low mutant allele frequency.
- Run dilutions for limit of detection.
- Perform reproducibility runs.
When assessing a new panel, Dr. Corless recommends considering the following: Are there areas with low coverage, and are known mutation hotspots within these regions? What is the lower limit of detection and how does DNA deamination affect this? What is the size range of insertions and deletions that can be detected?
Another critical factor in NGS testing on tumor samples is assessing the material that is being tested. At Knight, the lower limit of an acceptable sample is 20 percent tumor content. “As a general rule, pathologists tend to overestimate how much tumor they have in the starting material, and this is something we need to keep in mind,” Dr. Corless said.
The amount of tumor content in a sample is especially important when detecting copy number alterations, which are important in precision medicine. Knight has developed an algorithm to detect CNAs in amplicon-based sequencing data. The laboratory used samples with a known FISH status and microarray results to validate the results of the algorithm.
Ultimately, analytical validation of cancer panels should include normal samples, samples with a wide range of mutations, and samples with low mutant allele frequency, Dr. Corless advised. It is important to correlate sequencing results with tumor input, he said. Copy number alteration can be detected, but there are limitations. RNA sequencing also can be useful in detecting gene fusions.
Knight Diagnostic Laboratories has invested a lot to develop its own genomic database, but also relies on public resources such as My Cancer Genome (www.mycancergenome.org), along with the databases from MD Anderson Cancer Center (https://pct.mdanderson.org) and Washington University in St. Louis (https://civic.genome.wustl.edu).
Any laboratory venturing into NGS testing will need to have a robust genomic database, Dr. Corless noted. A laboratory might start with one that is commercially available and then adapt it to its own setting, or may invest in developing one of its own as Knight has done.
[hr]
Kim Scott is a writer in Lewes, Del.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
