Three rapid panels put to the DOOR-MAT test
Sherrie Rice
April 2022—A study published last fall examined antimicrobial prescribing in gram-negative bloodstream infections based on three rapid diagnostic panels and using what’s known as the DOOR-MAT framework.
The study’s findings (Claeys KC, et al. Clin Infect Dis. 2021;73[6]:1103–1106) were explained in a CAP TODAY webinar on stewardship interventions to optimize the management of gram-negative bacteremia. It was presented last December and made possible by a special educational grant from BioFire. (The full webinar is at captodayonline.com.)
The question is not whether rapid diagnostic tests should be used to aid in managing bloodstream infections, especially gram-negative BSIs, Kimberly Claeys, PharmD, associate professor of infectious diseases, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, said in the webinar. “Even given the challenges we face with them,” she said, “there is benefit to using them. The question then goes to which one we should be using to optimize the management of our gram-negative BSIs in our respective institutions.”

Dr. Claeys
In their proof-of-concept study, Dr. Claeys and colleagues compared three rapid diagnostic test panels—Verigene BC-GN (Luminex) and BioFire FilmArray BCID and BCID2 (the latter was RUO at the time)—using the Desirability of Outcome Ranking for the Management of Antimicrobial Therapy, or DOOR MAT. The aim was to evaluate potential downstream prescribing decisions resulting from the different organism and resistance mechanism detection of the panels.
“Most of the comparative literature doesn’t help us nail down our questions about prescribing patterns or patient outcomes,” Dr. Claeys said, “and we don’t get a sense of how one tool compares to another in a real-world environment.”
Their retrospective study was conducted from August 2018 to August 2019. Blood cultures from adult patients with gram-negative organisms identified via Gram stain were eligible for inclusion. (If gram-positives were also isolated, patients were excluded.) The Verigene BC-GN microarray panel was used as part of routine clinical practice during the study. The FilmArray BCID was run on fresh samples within eight hours of detection, and BCID2 was run on retained blood samples frozen at -80° C after routine testing. The FilmArray results weren’t made available to providers. Final organism identification and phenotypic susceptibility results were based on Vitek MS/Vitek 2 automated susceptibility testing.
The primary objective was to evaluate potential differences in antimicrobial prescribing based on panel results, using DOOR-MAT analysis, and the secondary objective was to compare the on-panel positive percent agreement between the platforms and Vitek 2.
“DOOR MAT is a way to help us compare these tools without having to do a full-blown, resource-intensive, randomized controlled trial,” Dr. Claeys explained, noting that before her work and that of her coauthors, DOOR MAT had not been applied to directly compare multiple rapid diagnostic panels. The method was developed by the Antibacterial Resistance Leadership Group, which was created in 2013 and is funded by the NIH National Institute of Allergy and Infectious Diseases and facilitated by the Duke Clinical Research Institute (Wilson BM, et al. Clin Infect Dis. 2021;73[2]:344–350).
“Through use of DOOR MAT, institutions can better determine which RDTs [rapid diagnostic tests] to implement based on local infectious diseases epidemiology and prescribing patterns,” she and coauthors write.
In their study, 108 gram-negative organisms were identified from 103 patients. The most common source of bloodstream infection was urinary (31 percent), followed by unknown (23.3 percent). In 43 percent of patients, the blood cultures were obtained in the ICU. “Not surprisingly, on-panel positive percent agreement between all three panels was high and very consistent,” Dr. Claeys said. For Verigene BC-GN, positive percent agreement was 98.8 percent; for FilmArray BCID, 97.8 percent; and for BCID2, 96.9 percent.
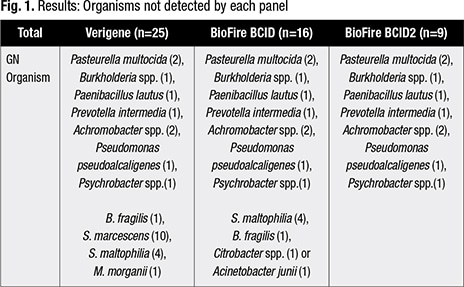
More interesting, she said, is what is not detected on each panel. Of the 108 organisms detected, Verigene didn’t detect 25 of them, BCID did not detect 16, and BCID2 did not detect nine (Fig. 1). “So the big take-home there is that the BCID2 detected 60 percent of the organisms not on the Verigene panel and 24 percent of the organisms not on the BCID panel.” (Verigene BC-GN and BCID2 accurately identified the six CTX-M, which was the only genetic resistance detected.)
The mean DOOR-MAT scores are as follows: Verigene BC-GN: 83.8 (SD ± 25.7); BCID: 59.9 (SD ± 33.7); and BCID2: 89.7 (SD ± 24.7). The higher mean score for BCID2, when compared with the Verigene panel, is attributed to the detection of an expanded panel organism that the BCID2 can detect, the authors write. “When compared with BioFire BCID,” they continue, “the significantly expanded detection of resistance determinants allows not only for prompt escalation, such as in the presence of CTX-M production, but also allows the potential to de-escalate” in its absence.
“We want to be able to choose the antimicrobial that’s most narrow in spectrum while still having in vitro activity, to allow for the best clinical outcomes, while exerting minimal selective pressure on future resistance,” Dr. Claeys said.
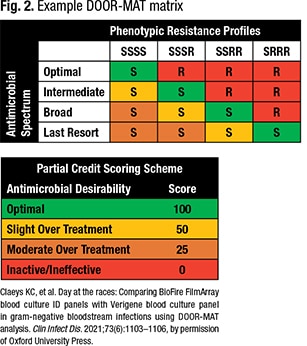
Step one to developing a DOOR-MAT score is to define the antimicrobials of interest and their spectrum of activity and to rank them from most narrow to most broad. Step two is to determine the resistance profiles for organisms of interest based on the antimicrobial spectrum levels. “So you go from an organism that is pan-susceptible all the way to an organism that is resistant to everything but the agent of last resort,” Dr. Claeys said. Step three is to create a matrix or framework that cross-references the spectrum of activity levels and resistance profiles. Step four is defining the partial credit scoring system to go with the framework (Fig. 2). Zero is the least desirable therapy, and “100 is your most narrow, effective therapy,” she said.
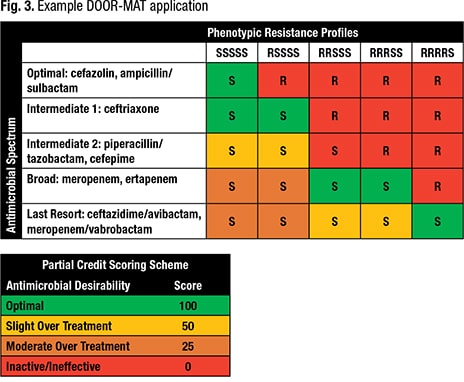
Dr. Claeys provided a sample clinical scenario: a 41-year-old female patient on a transplant medical floor (not ICU), gram-negative bacteremia 2/2 urinary source. No known drug allergies or prior multidrug resistances. Previous antibiotics include vancomycin and cefazolin. Results for the BC1D2 and Verigene panels are as follows: E. coli, CTX-M negative. For BCID: E. coli.
Given the patient information, the known local infectious diseases epidemiology, and the rapid diagnostic test results for each panel, the agents chosen could be ceftriaxone for E. coli, CTX-M negative, and piperacillin-tazobactam for E. coli but unknown CTX-M. “We might keep the patient on our workhorse agent and wait for final susceptibilities,” she said of the latter. And the susceptibility profile reveals the E. coli is a pan-susceptible organism (Fig. 3). In Fig. 3, “you can see, within the framework that we developed, ceftriaxone is going to be given an optimal score for E. coli in these situations, based on our partial credit scoring system.”
The Verigene and BCID2 have scores of 100. BCID has a score of 50 because “we are stuck with piperacillin-tazobactam because we weren’t sure about molecular resistance. It fits into our framework as intermediate category two, which we’ve defined as slight overtreatment.” If perhaps the patient had been in the ICU, or had a prior multidrug-resistant organism, “that would still go into this broad category, moderate overtreatment, and the score would be 25.”
That is how DOOR MAT works, Dr. Claeys said. “You just rinse and repeat throughout each case.”
To sum up, she said all platforms have a high level of agreement for on-panel targets, and BCID2 detected many organisms that were not on the other two panels. “And through DOOR MAT, we demonstrate that these in vitro differences may also have a clinical benefit in terms of antimicrobial prescribing decisions.”
For rapid diagnostic testing and antimicrobial stewardship, the focus of most of the data in the literature are the gram-positive organisms causing bloodstream infections, said J. Kristie Johnson, PhD, D(ABMM), a co-presenter in the webinar and professor in the University of Maryland School of Medicine Department of Pathology. Laboratories and antimicrobial stewardship teams have been slower to adopt gram-negative rapid diagnostic tests, and there are several reasons, she said, noting they are a little more difficult.
Dr. Johnson
One reason is there is more genetic diversity in the gram-negative organisms that are pathogenic. Another is that they have numerous and complex mechanisms of resistance. And with the current rapid diagnostic tests, “there is incomplete data, there are organisms that might not be on the assay, and resistance mechanisms that aren’t detected on the assay,” Dr. Johnson said. “And that makes some of the decisions for escalating or de-escalating more difficult.” In addition, polymicrobial infections with gram-negative bacteria can be hard to detect.
In the efforts to improve antimicrobial prescribing at the University of Maryland, the laboratory and pharmacy departments meet weekly, said Dr. Johnson, medical director of microbiology. “We discuss the priorities for both the lab and antimicrobial stewardship to understand the barriers both teams have and to work with those and move forward. In the end we both want the same outcome,” Dr. Johnson said.
The infectious disease physicians have been pleased, she said. The laboratory switched from the Verigene platform to BCID2, changed its reporting structure, and provided education. Previously, when using a rapid diagnostic test, the laboratory interpreted the result and reported it in the blood culture report. “Now we report out each organism if it was detected or not detected. Our infectious disease group knows the result will be there and looks for it,” Dr. Johnson said, “and we worked with them to make it easy for them to understand the results.” With Dr. Claeys and colleagues, the laboratory developed algorithms based on what the assays detect and what therapies should be considered.
Said Dr. Claeys, “Our infectious disease colleagues respect that we are helping them with these technologies and being a liaison with the microbiology laboratory.”
Sherrie Rice is editor of CAP TODAY.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
