Amy Carpenter Aquino
October 2021—Clonal hematopoiesis is a significant biological phenomenon and denotes presence of mutations in bone marrow stem cells in the absence of a hematologic malignancy. Prevalence rises with aging and certain therapeutic or environmental exposures. CH has been associated with increased risk for leukemias, cardiovascular disease, and mortality.

Dr. Razavi
A study presented by Pedram Razavi, MD, PhD, a medical oncologist and physician-scientist at Memorial Sloan Kettering Cancer Center, in an AMP session last year, revealed that CH is far more prevalent in patients with cancer as well as in healthy individuals than previously thought, making it a technical pitfall in cell-free DNA testing for detection of circulating tumor DNA (Razavi P, et al. Nat Med. 2019;25[12]:1928–1937).
Sequencing at high depth can identify the majority of the cfDNA clonal hematopoiesis mutations but not all.
In the study (Fig. 1), which used matched cfDNA-WBC sequencing, CH was not found to be limited to the older patients. “Almost all the cancer patients and the healthy individuals had some level of detectable CH in the white cells and in the cell-free DNA,” regardless of age, Dr. Razavi said. The number of CH variants in an individual was found to be associated with age, but it was highly variable, he said, “with many of the young patients also having high levels of CH.”
Dr. Razavi and colleagues sought to define the technical feasibility of a high-intensity sequencing assay of cfDNA and matched white blood cell DNA covering a large genomic region in their prospective study of 124 patients with metastatic cancer (with matched tumor tissue biopsies) and 47 noncancer controls. “At the time of progression or de novo metastatic diagnosis, we collected cell-free DNA and tissue at very short time intervals without any intervening therapy,” Dr. Razavi said.
The cfDNA and white blood cells were sequenced to a minimum target depth of 60,000×. “We spent a lot of time on the joint calling, noise canceling, and noise reduction modeling,” he said, using a machine-learning–based error model. For tumor sequencing, “we used our well-validated MSK-IMPACT assay that includes tumor and normal sequencing of 410 genes.”
A comparison of raw and collapsed sequence depth across the patients with cancer and the noncancer controls showed similar depth of sequencing, he said. The depth—about 70,000× for cfDNA and WBCs—“as expected, was associated with input DNA, with the input DNA being around 12 to 75,” and 75 the maximum DNA input for both, Dr. Razavi said.
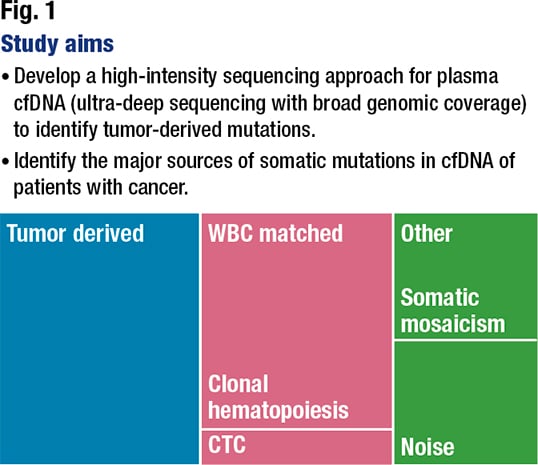
After implementing the error correction and joint cfDNA and WBC variant calling, the team categorized the cfDNA fragments into five variant groups. If they were germline, no further analysis was done. If they were biopsy matched, it meant they were found by MSK-IMPACT and reported in the clinical MSK-IMPACT report. The biopsy subthreshold category was for the mutations found in the binary alignment map files of the tumor MSK-IMPACT (≥ 3 reads) but that did not pass the threshold for clinical mutation calling. The WBC-matched mutations were those found in the white cells that were not germline or tumor-derived (biopsy matched or subthreshold). And variants of unknown source were none of the above and could not be matched.
“The assay showed a high sensitivity and a low false-positive rate,” with performance of the cfDNA assay comparable to that of Droplet Digital PCR, and high reproducibility in independent biological replicates, Dr. Razavi said.
“As expected, the majority of the tumor-derived mutations were found in cell-free DNA,” he said, “and we’ve been able in most of the cases to find at least one tumor-derived mutation in the cell-free DNA. And the patterns of mutation followed what we expect for the specific cancer type.”
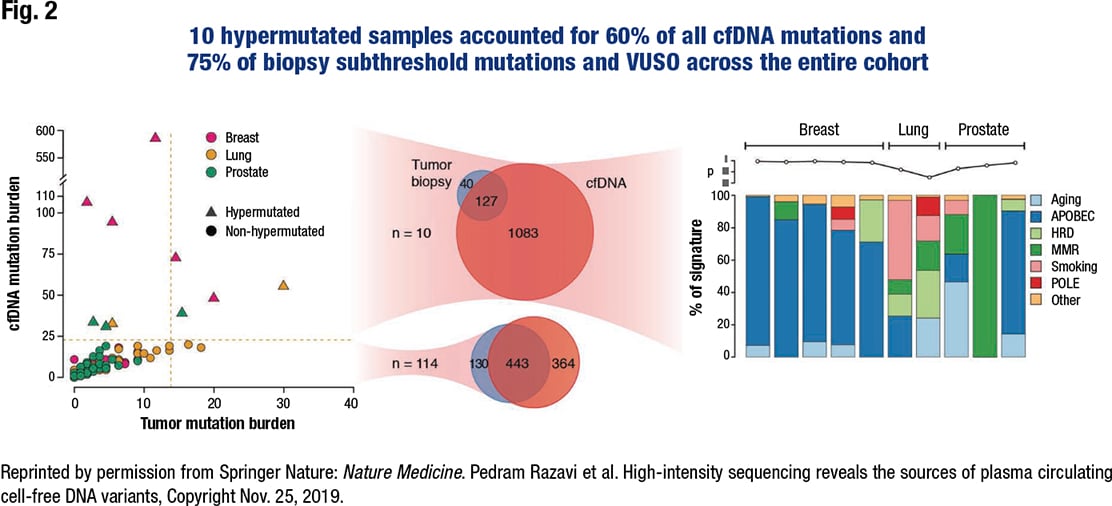
The surprise was that the 10 hypermutated samples accounted for 60 percent of all cfDNA mutations and 75 percent of biopsy subthreshold mutations and variants of unknown source across the entire cohort (Fig. 2). “In the other 114 patients,” Dr. Razavi said, “the match between the tumor and cell-free DNA was very good.”
He and his colleagues analyzed the mutational signature on the 10 hypermutated samples to show the utility of the assay for such an analysis. “We found what we expected in most of the breast cases” (n = 5): an APOBEC mutational signature, known to amplify tumor heterogeneity and subclonal diversity. One of the three hypermutated prostate cancer cases displayed the same; another had a dominant MMR signature. “In lung cases, it was either smoking or aging,” he said.
Another surprise: The vast majority of somatic cfDNA mutations in controls and nonhypermutated cancer patients were WBC matched.
“We expected most of the somatic mutations in the healthy individuals to come from the white blood cells and originate from clonal hematopoiesis,” Dr. Razavi said, “but in cancer patients we were surprised to see more than 50 percent of the mutations we found in nonhypermutated cases were also found in the white cells.”
The WBC-matched mutation burden was not associated with the tumor-matched mutational burden, “telling us this is probably not a sequencing noise,” Dr. Razavi said. “That prompted us to think that this is most likely coming from clonal hematopoiesis.”
Bone marrow stem cells accumulate mutations as they replicate; some mutations are passenger mutations and don’t result in clonal expansion but nevertheless exist in the stem cell and daughter population coming from that stem cell. Some mutations result in expansion and selection of the subclone. “In cancer patients, this is even more important because we impose bottlenecks by giving patients cytotoxic therapies,” Dr. Razavi said. Some of these mutations result in resistance to therapy and expansion of these subclones further on, after each therapy cycle. “That’s why we probably see more clonal hematopoiesis in cancer patients, and some of the treatments we give probably induce hematopoiesis in the bone marrow” (Bowman RL, et al. Cell Stem Cell. 2018;22[2]:157–170).
The frequency of clonal hematopoiesis increases with age and starts peaking at age 60 to 65, Dr. Razavi said. Traditional assays using whole exome sequencing with a lower depth of sequencing have been able to find clonal hematopoiesis at levels of five to 10 percent in the WBCs. MSK’s high-depth sequencing using the MSK-IMPACT assay showed clonal hematopoiesis can be found at the younger ages and in a larger proportion of patients, Dr. Razavi said.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
