CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from Quest Diagnostics. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Lisa M. Blazejewski, MS
Neng Chen, PhD
Ke Zhang, PhD
Franklin Quan, PhD
December 2020—Circulating cell-free DNA in the blood of pregnant women is derived from both maternal tissues and the placenta.1 As a result, cfDNA isolated from maternal plasma can be used for noninvasive prenatal screening (NIPS) to identify fetal autosomal aneuploidies (trisomies 13, 18, and 21) and sex chromosome aneuploidies (SCAs). For fetal autosomal aneuploidies, NIPS offers higher detection rates and lower false-positive rates than traditional screening methods, such as maternal serum screening and nuchal translucency.2 NIPS is the only screening option available for SCAs, such as Turner syndrome (45,X) and Klinefelter syndrome (47,XXY), which do not present with ambiguous genitalia on fetal ultrasound.3
NIPS assays use multiple techniques, including single-nucleotide polymorphism, chromosome-specific, and whole-genome-based analyses. Our laboratory performs NIPS via massively parallel shotgun sequencing (MPSS) of cfDNA from maternal plasma. After cfDNA isolation, sequencing-ready libraries are prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina as described.4 Sequence reads are aligned to unique “bins” on autosomes and sex chromosomes. The number of reads mapping to each bin is used to calculate a chromosome-specific Z-score using a proprietary in-house–developed bioinformatics pipeline. The Z-score is thus a quantitative measure that can be used to determine if the representation of chromosomal material is greater or less than expected for a diploid fetus. Since a female fetus typically has two X chromosomes, and a male fetus only one, the X chromosome Z-score for a female fetus is usually greater than that of a male fetus. Increased risk for fetal autosomal aneuploidy or SCA is identified when a Z-score for an autosome or the X chromosome is outside of an empirically established, validated range for a fetus with a normal chromosomal complement. Sex determination is performed by counting the number of sequence reads that map to Y chromosome bins.4
However, since NIPS based on cfDNA from maternal plasma is a quantitative analysis of both maternal and placental cfDNA, NIPS can be affected by a variety of maternal conditions.5 As illustrated by the case presented in this report, another factor that can affect NIPS is a fetal demise in a multiple gestation pregnancy.
Patient case. A 36-year-old woman with a reportedly singleton pregnancy had a blood specimen drawn for NIPS at 10 weeks, one day gestation. The results did not indicate an increased risk for autosomal aneuploidies (trisomies 13, 18, and 21), and the X and Y chromosome data were consistent with a male fetus (NIPS draw 1, Table 1).
A second-trimester ultrasound revealed no abnormalities, but the fetal sex was determined to be female. Because of the discordant fetal sex determinations between NIPS and ultrasound, the physician obtained a second specimen at 16 weeks, six days gestation for NIPS. The X and Y chromosome data for the second NIPS were again consistent with a normal male fetus (NIPS draw 2, Table 1). Since the laboratory was unaware that the ultrasound fetal sex determination was female, a report for a normal male fetus was issued. Consequently, the patient was offered diagnostic testing to assess the fetal karyotype; this testing was declined.
In a further attempt to resolve the discordant sex determinations, the physician drew a third specimen at 25 weeks, six days gestation for NIPS. Again, the results did not indicate an increased risk for an autosomal trisomy. However, the sex chromosome data were such that the fetal sex could not be predicted. The number of Y chromosome reads this time was lower than expected for a male fetus and only slightly greater than the range established as background for female fetuses. In addition, the X chromosome Z-score was in the range expected for a female fetus (NIPS draw 3, Run No. 1, Table 1). Repeat testing performed on this specimen yielded similar results (NIPS draw 3, Run No. 2, Table 1). These results were suggestive of either mosaicism for an SCA (e.g. 46XX/46XY mosaicism) or a vanishing twin (VT) pregnancy. A VT pregnancy is one in which a twin gestation is reduced to a singleton gestation after the demise of one fetus. In this case, the sex of the demised fetus would be predicted to be male.
A comparison of the sex chromosome data for all three specimens (Table 1) revealed that the number of Y chromosome sequence reads decreased significantly with advancing gestational age. They declined from a level clearly indicative of a male fetus at 10 weeks, one day to a level not much greater than that expected for a female fetus at 25 weeks, six days, with the number of reads at 16 weeks, six days less than that observed at 10 weeks, one day but still clearly indicative of a male fetus. In addition, the representation of X chromosome material increased over this time, from a level consistent with a male fetus to one consistent with a female fetus.
The trends in the sex chromosome data were consistent with a VT pregnancy with the demise of a male fetus and provided an explanation for the discordance between the NIPS and ultrasound fetal sex determinations. At each time point, the fetal cfDNA isolated from the maternal plasma included both cfDNA from a viable female fetus and cfDNA from a likely male demised fetus, with the contribution from the demised male fetus decreasing over time. The cfDNA from the demised male fetus was initially sufficient to cause the NIPS sex prediction to be unequivocally male. However, as male cfDNA from the demised twin was cleared from the maternal circulation, the NIPS sex prediction became equivocal and trended toward female.
To confirm a VT pregnancy, a laboratory genetic counselor contacted the physician for clinical information. At this time, all of the clinical background for the pregnancy was provided, including the fact that the pregnancy was originally a spontaneous twin gestation with the demise of one twin. The NIPS specimen drawn at 10 weeks, one day gestation was drawn immediately after the recognition of that demise. The female ultrasound sex determination was also provided. Postnatal follow-up confirmed delivery of a female baby.
Discussion. NIPS performed using cfDNA from maternal plasma is an effective screening tool for the detection of fetal autosomal aneuploidies and SCAs and the determination of fetal sex. NIPS offers higher detection rates and lower false-positive rates for autosomal aneuploidy screening than do traditional methods and is the only screening option available for SCAs. However, as illustrated by the case described here, NIPS predictions of fetal sex can be confounded in the setting of a VT pregnancy. In addition, VT pregnancies involving the demise of a twin with an autosomal aneuploidy can lead to false-positive NIPS results.6,7
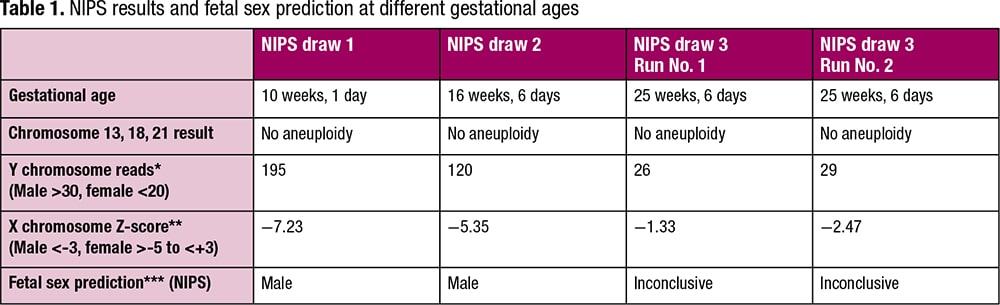
*The sex of the fetus is predicted to be male when the number of reads mapping to the Y chromosome is greater than 30 and female when the number of reads mapping to the Y chromosome is less than 20.
**For a male fetus the X chromosome Z-score is <-3, and for a female fetus the X chromosome Z-score is >-5 and <+3.
***“Inconclusive” indicates the fetal sex could not be predicted from the NIPS data.
In a VT pregnancy, cfDNA from the demised twin can persist in the maternal circulation for a period of up to eight to 15 weeks after the fetal demise.8,9 This can cause NIPS results for fetal sex or autosomal aneuploidies in a known VT pregnancy to be discordant with ultrasound findings or the results of other screening or diagnostic testing. Conversely, since VT pregnancies may not always present via ultrasound, a VT pregnancy should be considered when NIPS results for an apparently singleton pregnancy are discordant with the results of other diagnostic testing. Since VT pregnancies are at higher risk for adverse maternal and neonatal outcomes,10 such as gestational diabetes, chronic hypertension, intrauterine growth restriction, and preterm labor, by alerting providers to the possibility of a VT pregnancy, NIPS could contribute valuable information to providers and potentially allow fetal and obstetric risks to be managed more successfully.
In addition, for appropriate interpretation of NIPS, it is essential that ordering physicians provide all relevant clinical information, such as a pregnancy being a VT pregnancy, to the testing laboratory. For this case, the report of a VT pregnancy would have provided an explanation for the discordance between the NIPS and ultrasound fetal sex determinations and would have eliminated the need for multiple rounds of NIPS.
- Liao GJW, Chiu RW, Lo YM. Prenatal assessment of fetal chromosomal and genetic disorders through maternal plasma DNA analysis. Pathology. 2012;44(2):69–72.
- Bianchi DW, Parker RL, Wentworth J, et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808.
- McKinlay Gardner RJ, Amor DJ. Chromosomal Disorders of Sex Development. In: Gardner and Sutherland’s Chromosome Abnormalities and Genetic Counseling. 5th ed. Oxford University Press; 2018:535–543.
- Strom CM, Anderson B, Tsao D, et al. Improving the positive predictive value of non-invasive prenatal screening (NIPS). PLoS ONE. 2017;12(3):e0167130.
- Bianchi DW. Cherchez la femme: maternal incidental findings can explain discordant prenatal cell-free DNA sequencing results. Genet Med. 2018;20(9):910–917.
- Grömminger S, Yagmur E, Erkan S, et al. Fetal aneuploidy detection by cell-free DNA sequencing for multiple pregnancies and quality issues with vanishing twins. J Clin Med. 2014;3(3):679–692.
- Hochstenbach R, Elferink MG, van Zon PHA, et al. Discordant NIPT result in a viable trisomy-21 pregnancy due to prolonged contribution to cfDNA by a demised trisomy-14 cotwin. Clin Case Rep. 2018;6(5):788–791.
- Sylvester-Armstrong KR, Rasmussen SA, Shoraka M, et al. Fetal sex discordance between noninvasive prenatal screening results and sonography: a single institution’s experience and review of the literature. Birth Defects Res. 2020;112(4):339–349.
- Niles KM, Murji A, Chitayat D. Prolonged duration of persistent cell-free fetal DNA from vanishing twin. Ultrasound Obstet Gynecol. 2018;52(4):547–548.
- Evron E, Sheiner E, Friger M, Sergienko R, Harlev A. Vanishing twin syndrome: is it associated with adverse perinatal outcome? Fertil Steril. 2015;103(5):1209–1214.
Lisa Blazejewski is a certified genetic counselor, Dr. Chen and Dr. Quan are senior directors, and Dr. Zhang is principal scientist—all at Quest Diagnostics.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
