CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from Ohio State University College of Medicine. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Matthew R. Avenarius, PhD
Lynne V. Abruzzo, MD, PhD
June 2020—A middle-aged adult presented with shortness of breath and bruising and was found to have leukopenia (WBC 4.16 K/mcL; normal range 4.50–11.00 K/mcL) and anemia (Hgb 7.6 g/dL; normal range 12–16 g/dL) with rouleaux. The hypercellular bone marrow core biopsy (80–90 percent cellularity) contained 90 percent plasma cells of variable morphology, some with prominent nucleoli and occasional binucleate forms. Serum protein electrophoresis, immunoglobulin quantification, and immunofixation demonstrated an IgA kappa monoclonal gammopathy (6,163 mg/dL; normal range 84–381 mg/dL). Radiographic bone survey was normal.
The patient’s cytogenetic evaluation included a plasma cell myeloma fluorescence in situ hybridization (FISH) panel performed on magnetically separated CD138-positive cells. The panel interrogated loci of known diagnostic and prognostic significance in plasma cell myeloma including the following: CDKN2C(1p32.3)/CKS1B(1q21.3); BCL6 break-apart (3q27); CSF1R(5q33-34)/D5S23:D5S721(5p15.2); MYC break-apart (8q24); ATM(11q22.3)/TP53(17p13.1); ETV6 (12p13)/RUNX1(21q22); RB1(13q14)/LAMP1(13q34); CCND1(11q13)/IGH(14q32.3). Among these probe sets the patient was positive for three CKS1B signals (red), a finding that deviates from the normal CKS1B signal pattern (two red signals) and was interpreted as a gain of 1q (5.2 percent, cutoff <2.3 percent). Additionally, the RB1 (red) and LAMP1 (green) probes each produced a single signal in the patient sample (normal controls demonstrated two red RB1 signals and two green LAMP1 signals), which is consistent with the loss of 13q (56.3 percent, cutoff <1.5 percent). Finally, multiple IGH/CCND1 fusion signals (six to 11 fusion signals per cell, 60.6 percent, cutoff <0.6 percent) were identified in the patient sample that deviated from the normal two red/two green pattern in controls indicating the amplification of the fusion product (Fig. 1). Normal signal patterns were observed for the remaining probes and were consistent with a near-diploid karyotype. Conventional cytogenetic analysis was unsuccessful. The patient received combination chemotherapy with carfilzomib, lenalidomide, and dexamethasone, followed by autologous stem cell transplant. The patient is alive 12 months after diagnosis with persistent disease.
The t(11;14)(q13;q32) is a recurrent chromosomal abnormality in plasma cell myeloma and mantle cell lymphoma. This translocation places the cyclin D1 (CCND1) gene immediately downstream of the immunoglobulin heavy chain (IGH) enhancer and results in overexpression of CCND1.1 The CCND1 gene encodes cyclin D1, which is required for normal progression through the G1 phase of the cell cycle. Ectopically expressed cyclin D1 aberrantly complexes with cyclin-dependent kinase 4 (CDK4) and hyperphosphorylates RB1. In this inactive state, RB1 loses its ability to suppress the progression to S-phase and leads to cell cycle dysregulation. Additionally, increased levels of the cyclin D1-CDK4 complexes sequester p27 from the cyclin E-CDK2 complex, which also inactivates RB1 and further dysregulates the cell cycle. Either directly or indirectly, overexpression of cyclin D1 inhibits normal RB1 function and drives oncogenesis.
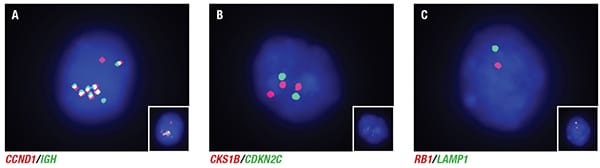
Fig. 1. Interphase nuclei demonstrating abnormal signal patterns for a subset of plasma cell myeloma probes. A. Enhanced micrograph of interphase nuclei probed with a dual-color/dual-fusion probe set that hybridizes to CCND1 and IGH. 60.6 percent (cutoff <0.6 percent) of cells counted demonstrate multiple fusion signals. B. Enhanced micrograph of interphase nuclei probed with CKS1B and CDKN2C. 5.2 percent (cutoff <2.3 percent) of cells counted were positive for a gain of 1q. C. Enhanced micrograph of interphase nuclei probed with RB1 and LAMP1. 56.3 percent (cutoff <1.5 percent) of cells counted were positive for a loss of 13q. The inset photomicrographs are unenhanced images.
Three reports of IGH/CCND1 fusion amplification are presented in the literature, two patients with mantle cell lymphoma and one patient with plasma cell myeloma. The first patient was a 58-year-old woman with mantle cell lymphoma.2 Initial cytogenetic analysis demonstrated multiple IGH/CCND1 fusion signals and hemizygous deletion of TP53 by interphase FISH analysis. Additional studies revealed a complex karyotype with multiple marker chromosomes, and metaphase FISH demonstrated IGH/CCND1 fusion signals on the der(2) and der(14) chromosomes. The patient died one month after diagnosis.
The second patient with mantle cell lymphoma was a 78-year-old man whose diagnostic bone marrow specimen revealed a complex karyotype including the t(11;14)(q13;q32).3 Metaphase FISH analysis demonstrated that the der(14) harbored a homogeneously staining region that was positive for amplification of the IGH/CCND1 fusion. Despite aggressive chemotherapy, the patient died of disease approximately 1.25 years after diagnosis.
The third patient was a 64-year-old man who presented with plasma cell leukemia.4 Chromosome analysis on bone marrow demonstrated a complex karyotype without evidence of a t(11;14)(q13;q32). However, interphase FISH analysis identified five IGH/CCND1 fusion signals in hypodiploid cells, and 10 IGH/CCND1 fusion signals in hypotetraploid cells. Despite aggressive multiagent chemotherapy, the patient died of progressive disease.
We report the fourth case of IGH/CCND1 fusion amplification, the second in a patient with plasma cell myeloma. While conventional cytogenetic analysis was unsuccessful, the presence of a 1q gain, 13q deletion, and IGH/CCND1 fusion amplification is consistent with a complex karyotype. Because reports of IGH/CCND1 amplification are few, it is difficult to determine the prognostic significance of this finding. However, its association with karyotypic complexity and the unfavorable clinical outcomes in the published cases suggest that this abnormality may portend a poor prognosis.
- Vogt N, Dai B, Erdmann T, Berdel WE, Lenz G. The molecular pathogenesis of mantle cell lymphoma. Leuk Lymphoma. 2017;58(7):1530–1537.
- Gruszka-Westwood AM, Atkinson S, Summersgill BM, et al. Unusual case of leukemic mantle cell lymphoma with amplified CCND1/IGH fusion gene. Genes Chromosomes Cancer. 2002;33(2):206–212.
- Miao Y, Lin P, Wang W, Jeffrey Medeiros L, Lu X. CCND1-IGH fusion-amplification and MYC copy number gain in a case of pleomorphic variant mantle cell lymphoma. Am J Clin Pathol. 2016;146(6):747–752.
- Ishigaki T, Sasaki K, Watanabe K, et al. Amplification of IGH/CCND1 fusion gene in a primary plasma cell leukemia case. Cancer Genet Cytogenet. 2010;201(1):62–65.
- Plasma Cell Neoplasms (Including Multiple Myeloma) Treatment (PDQ)—Health Professional Version. National Cancer Institute website. www.cancer.gov/types/myeloma/hp/myeloma-treatment-pdq. Updated July 19, 2019.
Dr. Avenarius is assistant professor, Department of Pathology, and associate director, molecular pathology laboratory, Department of Pathology, Ohio State University, Columbus. Dr. Abruzzo is professor and medical director, clinical cytogenetics laboratory, Department of Pathology, Ohio State University.
Test yourself
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
