Discussion. The decision to screen with either IHC, PCR, or both assays requires careful consideration of the technical aspects and limitations of each method.9,10 IHC is an attractive screening assay because it is relatively inexpensive and simple to perform in most pathology laboratories. IHC can be performed on small biopsies composed entirely of tumor tissue. The IHC staining pattern may also suggest which gene (MLH1, MSH2, MSH6, or PMS2) is affected, allowing for efficient triaging of further analysis. A problematic aspect of IHC testing is that staining may vary in intensity or lack convincing positive internal controls, and interpretation requires an experienced pathologist. The greatest limitation of IHC is that not all pathogenic MMR mutations result in loss of immunoreactivity, such as missense mutations or frameshift/truncation mutations that interfere with protein function without altering the antigenic portion of the protein. A study by Bartley, et al., determined that 11.8 percent of MSI-H carcinomas retained IHC expression of all four MMR proteins, yielding a false normal IHC result.11 Thus, a screening strategy using solely IHC will miss a significant proportion of MSI-H tumors.
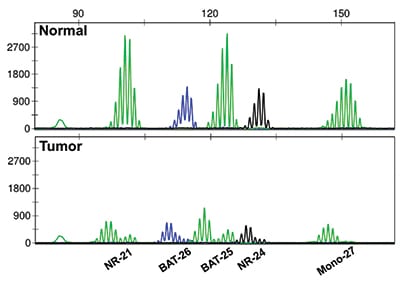
Fig. 2. PCR-based analysis of microsatellite length demonstrates microsatellite instability. Five mononucleotide repeat loci are assessed, comparing normal tissue (top panel) to tumor tissue (bottom panel).
PCR-based MSI assays can identify tumors with defective MMR regardless of MMR protein status. While the majority of germline MMR mutations occur within the proteins MLH1, MSH2, MSH6, and PMS2, the MMR machinery relies on myriad other genes and gene products such as MSH3, PMS1, or EPCAM, which can also cause MMR deficiency when mutated.10 Such mutations would be successfully identified by PCR, but not IHC. PCR assays are comparatively simple to evaluate and less susceptible to interpretation errors. Indeed, studies have shown that the reproducibility of PCR-based MSI testing is close to 100 percent.10 However, the assay must be performed in a more specialized laboratory, and the turnaround time is typically longer than that of IHC. PCR also requires both normal and tumor tissue for comparison, which may not be present in all small tissue biopsies, thus necessitating surgical resection for complete assessment. Unlike IHC, PCR results provide no clues as to which gene may contain the underlying mutation, thus offering no cost savings in triaging further analysis. Another important caveat is mutation of MSH6, which, due to functional redundancy in the MMR system, may or may not cause MSI that is detectable by PCR. A screening strategy that uses only PCR would identify such a tumor as MSS, a false normal result. Thus, neither IHC nor PCR is equipped to identify 100 percent of tumors with defective MMR, a problem that may be solved by next-generation sequencing, which can simultaneously detect MSI and screen for mutations in MMR genes. Currently, NGS is cost prohibitive as a screening strategy, but as costs continue to fall, NGS may supersede IHC and PCR as the favored test, as a recent study showed NGS assays to have sensitivity and specificity approaching 100 percent.12,13
Screening strategies differ by institution and may vary within the institution depending on the level of clinical suspicion for Lynch syndrome. Numerous studies have concluded that IHC and PCR have comparable sensitivity and specificity in detecting defective MMR.4,9,10 Although the NCCN recommends using only one test initially, the panel also acknowledges that when clinical suspicion for Lynch syndrome is high, a normal result via one method may warrant testing via a second method.8 At our institution, we favor concurrent initiation of IHC and PCR to minimize the patient impact of rare false normal results, despite the increased cost of redundant testing. However, this strategy can (albeit rarely) yield discordant results, as in this case. The aforementioned study by Bartley, et al., sought to quantitate the frequency of clinically significant discordances at the authors’ own institution over an eight-year period and identified IHC/PCR disagreement in 13 of 591 (2.2 percent) colorectal tumors.11 The majority of discordant tumors showed normal IHC with MSI-H PCR. A subset of these cases was ultimately determined to have germline alterations in MSH2, MLH1, or PMS2, reinforcing the concept of nonfunctional MMR proteins with retained antigenicity. The remaining cases remained unresolved, but possible explanations include defects in one of the lesser-known accessory MMR proteins. The converse discordance is also possible: abnormal IHC with MSS PCR. Possible explanations for this scenario include contamination of the PCR reaction with excess normal cells, loss of MSH6 IHC staining in the setting of prior chemotherapy or radiotherapy, or retained mismatch repair function in patients with germline MSH6 mutations.14 When discordant IHC and PCR test results occur, the tumor should be considered defective in MMR, and patients should be referred to genetic counseling for consideration of germline testing.
Summary. Given the implications for disease prognosis, surveillance, and predicted responses to therapy, it is recommended that all patients with newly diagnosed colorectal carcinoma undergo testing for MMR status. MSI testing by PCR and MMR protein assessment by IHC are highly concordant, but neither test alone is sufficient to capture 100 percent of tumors with defective MMR, and therefore dual PCR and IHC testing is an acceptable practice. Discordant IHC and PCR results, recently estimated to occur in 2.2 percent of colorectal tumors, most commonly reflect mutated nonfunctional MMR proteins with retained antigenicity.11 Discordant results should be interpreted as defective MMR, and additional testing should be performed to rule out a germline mutation.
- Ryan E, Sheahan K, Creavin B, Mohan HM, Winter DC. The current value of determining the mismatch repair status of colorectal cancer: a rationale for routine testing. Crit Rev Oncol Hematol. 2017;116:38–57.
- Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline summary from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J Oncol Pract. 2017;13(5):333–337.
- Bellizzi AM, Frankel WL. Colorectal cancer due to deficiency in DNA mismatch repair function: a review. Adv Anat Pathol. 2009;16(6):405–417.
- Geiersbach KB, Samowitz WS. Microsatellite instability and colorectal cancer. Arch Pathol Lab Med. 2011;135(10):1269–1277.
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–618.
- Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16(7):30.
- Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520.
- Provenzale D, Gupta S, Ahnen DJ, et al. NCCN Clinical Practice Guidelines in Oncology: Genetic/Familial High-Risk Assessment: Colorectal. Version 1.2018. https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf. Accessed Nov. 25, 2018.
- Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10(4):293–300.
- Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J Mol Diagn. 2008:10(4):301–307.
- Bartley AN, Luthra R, Saraiya DS, Urbauer DL, Broaddus RR. Identification of cancer patients with Lynch syndrome: clinically significant discordances and problems in tissue-based mismatch repair testing. Cancer Prev Res (Phila). 2012;5(2):320–327.
- Stadler ZK, Battaglin F, Middha S, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34(18):2141–2147.
- Nowak JA, Yurgelun MB, Bruce JL, et al. Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn. 2017;19(1):84–91.
- Gao Z. Pathology Review & Practice Guide, 2nd ed. Edmonton, Canada: Brush Education; 2017:744.
Dr. McCracken is a resident physician and Dr. Neff is an assistant professor in the Department of Pathology, Duke University Medical Center, Durham, NC. The authors thank Steven Conlon and Alexandra Scott for their work with image production and processing.
Test yourself
Here are three questions taken from the case report. Answers are online now at www.amp.org/casereports and will be published next month in CAP TODAY.
1. A recent report by Bartley, et al.,11 estimates the rate of discordance between IHC and PCR-based assays for mismatch repair at approximately:
a) 0.1 percent
b) 2 percent
c) 10 percent
d) 20 percent
e) 55 percent
2. Why is it important to test for mismatch repair/microsatellite instability status of colorectal cancers?
a) It provides key information about tumor staging.
b) Patients with intact MMR have a better overall prognosis.
c) Patients with intact MMR should have routine extra-colonic cancer screenings.
d) It provides potential risk information for patients’ family members.
3. Which of the following is the correct interpretation of a tumor’s mismatch repair status with intact MMR protein expression by IHC but MSI-high by PCR?
a) Intact
b) Deficient
c) Indeterminate
Test yourself answers
In the November 2018 issue was a case report, “Detection of concurrent hematologic malignancies in solid tumor NGS testing may cause false-positive results,” written by members of the Association for Molecular Pathology. Here are answers (in bold) to the three “test yourself ” questions that followed that case report.
1. KRAS mutations in lung adenocarcinoma are most likely to be found in which codon(s)?
a) 12 and 13
b) 13 and 61
c) 12 and 61
d) 61
2. KRAS mutations are usually mutually exclusive of which other mutations?
a) BRAF
b) EGFR
c) ALK
d) All of the above
3. RAS mutations are found in approximately what percentage of CMML?
a) 50 – 60 percent
b) 20 – 30 percent
c) 5 –10 percent
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
