Sherrie Rice
August 2020—The CAP treats emergency use authorization assays similar to FDA-cleared assays and thus requires full verification. In a June 4 CAP webinar, Neil Anderson, MD, D(ABMM), assistant director of clinical microbiology, Washington University School of Medicine in St. Louis, walked through how to approach verification for SARS-CoV-2 assays.
Co-presenter Elitza Theel, PhD, D(ABMM), director of the infectious diseases serology laboratory at Mayo Clinic, reported what’s known about protective immunity against SARS-CoV-2.

Dr. Anderson
For analytical interference, typical substances to investigate are hemoglobin, bilirubin, and triglycerides, and this should be performed at the limit of detection. “You may want to consider other exogenous inhibitors as well,” Dr. Anderson said, “maybe things you commonly see in samples submitted to your laboratory.” The laboratory can use data from the manufacturer in lieu of performing its own study.
Precision—the closeness of agreement between independent test measures—consists of intraassay (measurements collected under similar conditions) and interassay (under different conditions) precision. “Typical sources of imprecision need to be accounted for,” Dr. Anderson said, “and these include differences in timing of testing, temperature, mixing, pipetting, “pretty much anything you can introduce that might lead to a different result.” And it’s ideal to test concentrations at or near the limit of detection.
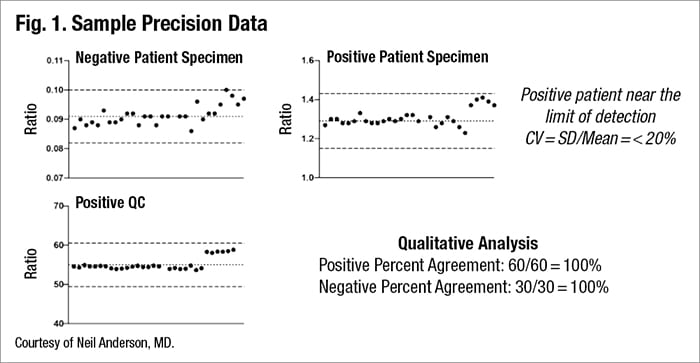
In his laboratory at Barnes Jewish Hospital, he and colleagues used a negative and a positive patient specimen and a positive QC specimen, and then compared the ratio between calibrator and signal intensity as seen in Fig. 1.

Dr. Theel
“What you can see is we have different results. We ran those single specimens using multiple replicates and were able to learn quite a bit. There is a spread of results,” he said. The lab wants to have an acceptable range for results when looking at this quantitatively, he said, and it’s often defined by CV (SD/mean), requiring that it remain below 20 percent.
Looking at the data qualitatively is also important. This allows calculation of positive percent agreement and negative percent agreement, “based on what the obtained results were expected to have been. Ideally this should be at or near 100 percent.”
Determination of reportable range typically doesn’t apply to SARS-CoV-2 assays at this time because all are designated as qualitative, but it must be determined if a lab reports results quantitatively.
“You’re probably going to spend the bulk of your assay verification in trying to determine accuracy, the extent to which a particular test is in agreement with a reference method or comparator,” Dr. Anderson said. At this point, the ideal comparators are specimens from patients with known SARS-CoV-2 infection, established with molecular testing and collected at a known time post-symptom onset. “We all know molecular testing might be imperfect so there could be debate as to whether this is ideal, but that’s what we’re left with right now,” he said. A secondary comparator would be specimens with known positive and negative antibody status tested using another validated or verified antibody test.
Comparing the reference method, if it’s a gold standard, with the method being evaluated is used to determine sensitivity and specificity. If the reference method isn’t a gold standard, it has to be phrased as positive or negative agreement. How the lab defines the reference method for accuracy studies affects the results. “So particularly if you’re using serum from positive patients, you need to consider the timing of serum collection.
“For instance, if I’m using serum that had been collected from symptomatic patients near the time of symptom onset, my assay is going to look like it performs quite poorly because we know serology in general is going to have poor sensitivity early in disease. However, if I give my serologic assay a chance by looking at specimens that have been collected 14 days or later from symptom onset, using that type of analysis I’m going to show a better sensitivity.”
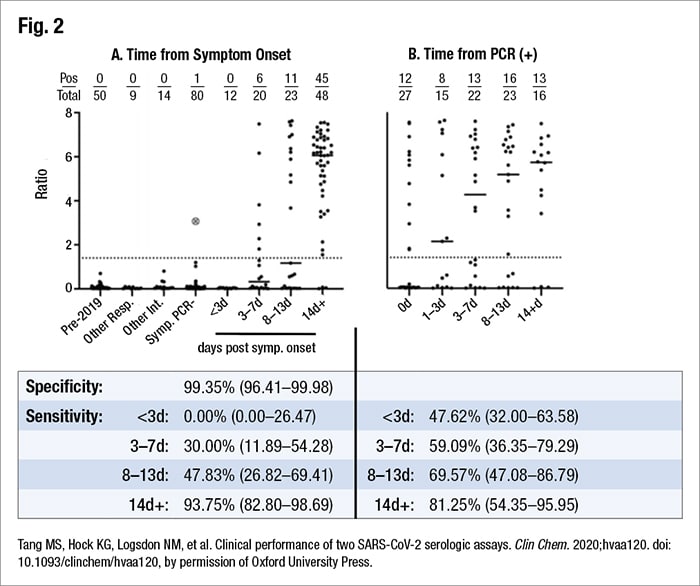
A lab might want to investigate both pieces of data, he argues: sensitivity early and late in disease. That’s how sensitivity was determined in his laboratory. They had 89 “positive” samples, based on PCR as a comparator. These were serum drawn at a variety of times post-symptom onset. They used remnant CBC samples from positive patients, with testing performed on the Abbott Architect.
“And what we found was an overall sensitivity of 56 percent. You might say that’s pretty abysmal, but when you look at the data and analyze it at different time points, the story becomes a little clearer,” Dr. Anderson said. “We did have low sensitivity at early onset of disease. However, once you get beyond 14 days, we had a 94 percent sensitivity”—sufficiently sensitive to put the assay into use.
Sensitivity varies based on how the data are analyzed, he said, pointing to his laboratory’s data, which were analyzed in two ways (Fig. 2). When they looked at the time from symptom onset, they saw the expected low sensitivity early in disease. When the lab looked at time from PCR, the data looks different. “What’s going to happen here is not everyone is going to necessarily get that PCR on their first day of symptomatology. So this is going to overestimate sensitivity early in disease.” Keep that in mind, he advises, and keep in mind, too, that “a lot of manufacturers are defining their sensitivity that way likely because all they have is the PCR data and they may not be able to do that chart review to figure out exactly when the patient became symptomatic.”
For an evaluation of specificity, formal and exhaustive cross-reactivity studies aren’t needed for an EUA assay, but accuracy studies should take into account common cross-reacting targets. “What I mean by that is you should try to include samples from patients with documented seasonal coronavirus positivity, with disease processes similar to COVID-19, so other respiratory diseases, and with common conditions that can lead to cross-reacting antibodies such as lupus or infectious mononucleosis. If these lead to antibodies that are going to react, that’s important information to know and communicate to your providers.”
Dr. Anderson calls cross-reactivity with seasonal coronaviruses “a real phenomenon,” as reported in the literature. “SARS-CoV-2 has a pretty high amino acid homology with SARS, less so with seasonal coronaviruses, though there is some homology there. So depending on the assay design, you may or may not see cross-reactivity.” Some studies have shown high cross-reactivities; others have shown very little, he said. “The bottom line, though, is that it’s theoretically possible, and to compound this, seroprevalence studies have suggested that a lot of us do have antibodies against seasonal coronaviruses. Sixty-five to 75 percent of young kids have antibodies to at least one seasonal coronavirus, and greater than 90 percent of adults older than 50 years of age have antibodies to all four coronaviruses.”
On the FDA website is information about the seasonal coronaviruses included in a manufacturer’s evaluation. It varies from no seasonal coronaviruses included to as many as 40, as of early June. “Most of them show no cross-reactivity, though I think I would argue that based on the amount of specimens tested even by some of the more thorough manufacturers, it doesn’t really capture the entire risk,” Dr. Anderson said.
He shared an example of a specificity study performed in his laboratory (using remnant CBC specimens). He and colleagues studied 110 “negative” samples: 50 were pre-COVID-19 outbreak, nine were from patients with other respiratory illnesses, and 14 were from patients with viral infections or other possible interferents. They found an overall specificity of 100 percent.
 CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
CAP TODAY Pathology/Laboratory Medicine/Laboratory Management
